BCMB 412: Transposition
1/43
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
44 Terms
The varigated phenotype in maize is a result of _______. Who discovered this?
transposition; Barbara McClintock
What is transposition? What is often a consequence of transposition?
A specific form of genetic recombination that moves certain genetic elements from one DNA site to another within the same genome. Transposition is a major source of mutation.
What is the RNA that represses expression of transposable elements?
piwi RNA
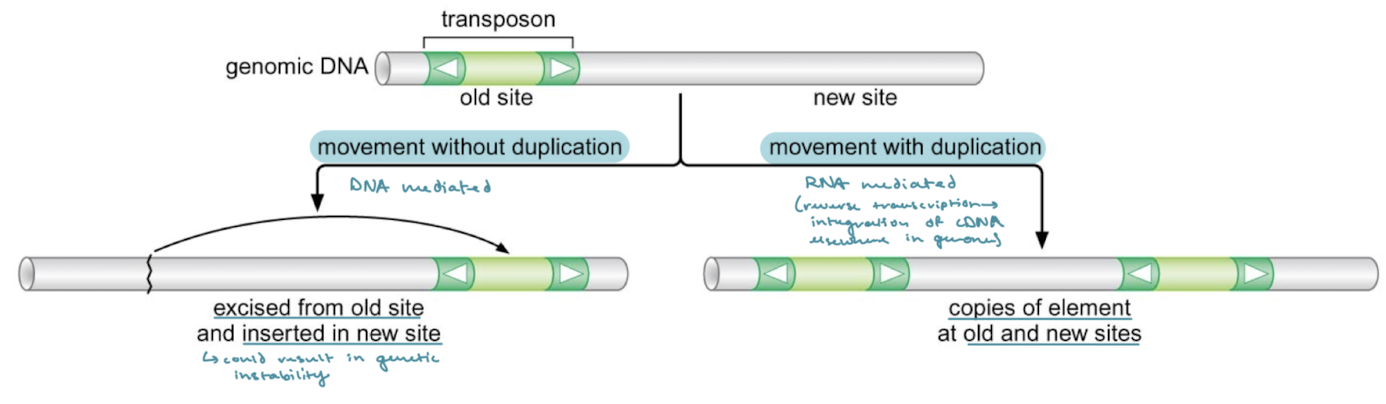
What are the two kinds of transposition? Which is DNA mediated and which is RNA mediated?
Movement without duplication: the transposon is excised from the old site and inserted into the new, which could result in genomic instability. This is a DNA-mediated process.
Movement with duplication: There are copies of the element at the old and new site. This is an RNA-mediated process because reverse transcriptase creates the cDNA that is integrated into the genome at the new site.
Put the different organisms in order of least to most occurrence of transposons: drosophila, s. cerevisiae, maize, E. coli, humans. What correlation does this order represent.
E. coli —> yeast —> drosophila —> human —> maize
There is a correlation between the size of the genome and the occurance of transposable elements.
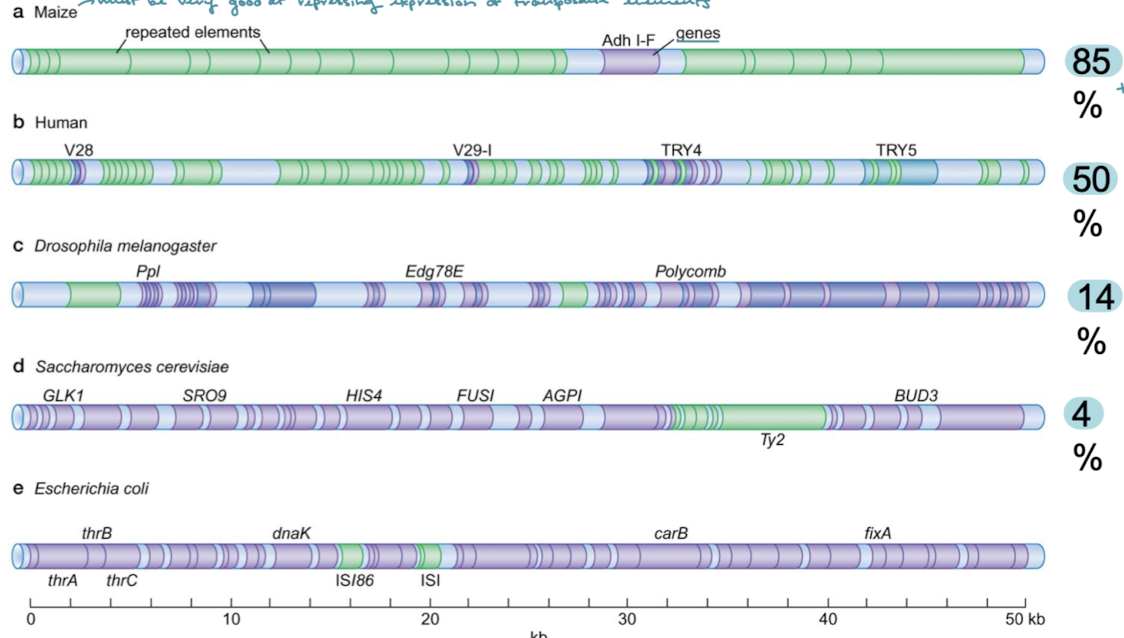
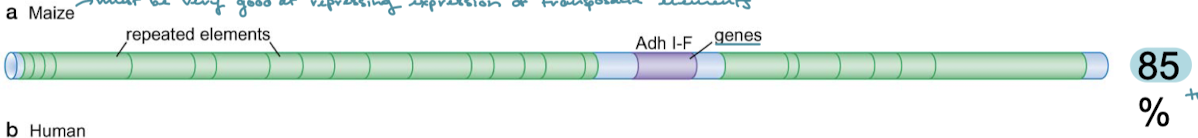
Maize has 80% transposable elements in its genome. Maize must be very good at what?
Repressing expression of transposable elements.
Retrotransposons have what kind of intermediates?
RNA intermediates
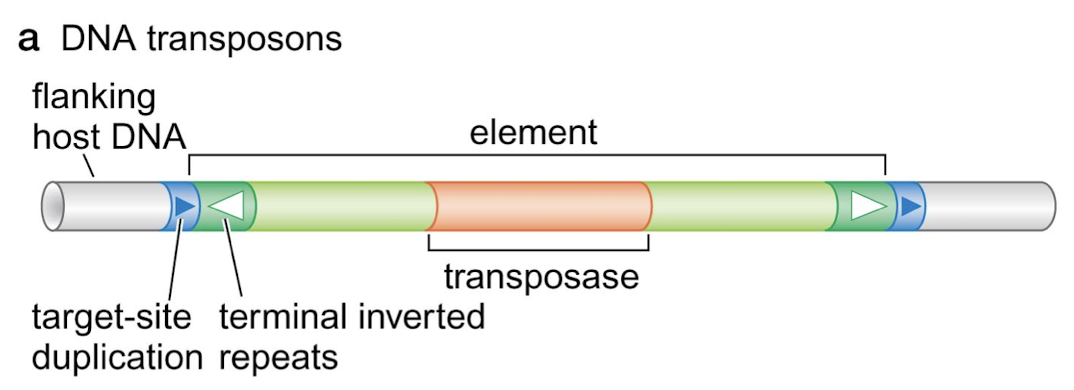
What are the three principal classes of transposable elements?
DNA transposons
Virus-like retrotransposons/retroviruses
Poly-A retrotransposons
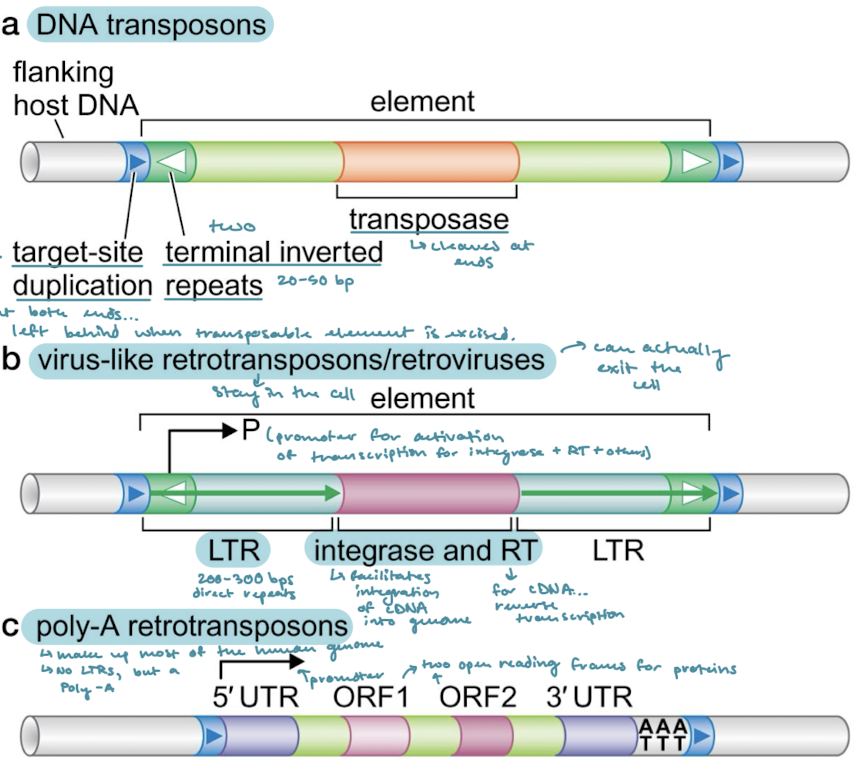
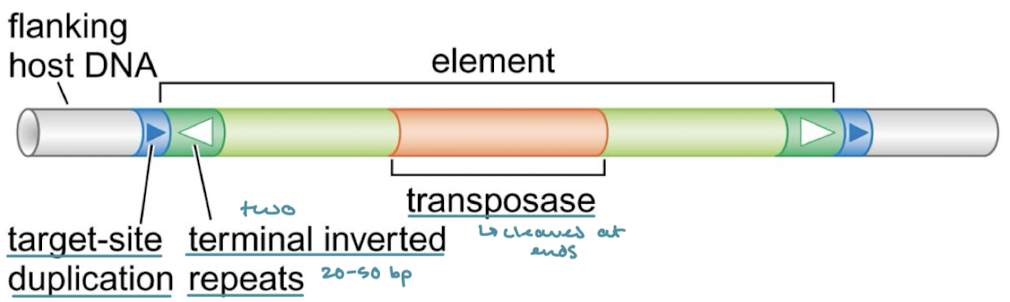
Which of the three principal classes of transposable elements have target-site duplications (direct terminal repeats). What are they?
They are common to all classes. They are like a footprint of a few nucleotides at both ends of the element that are left behind when a transposable element is excised.
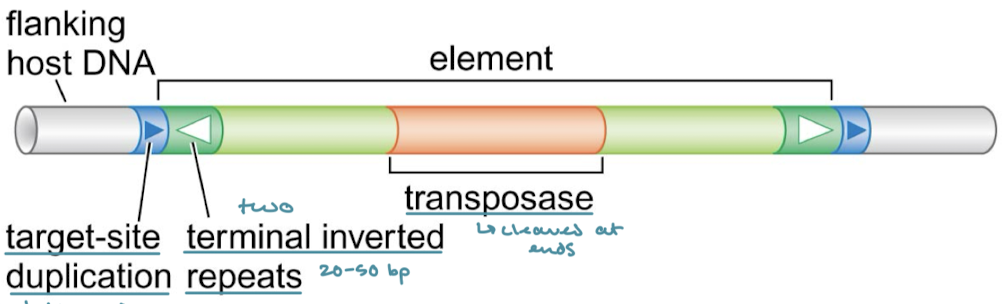
Which of the classes of transposable elements have terminal inverted repeats?
DNA transposons and virus-like retrotransposons/retroviruses
What is the protein that DNA transposons encode for within the element?
Transposase, responsible for cleaving the ends of the element for excision.
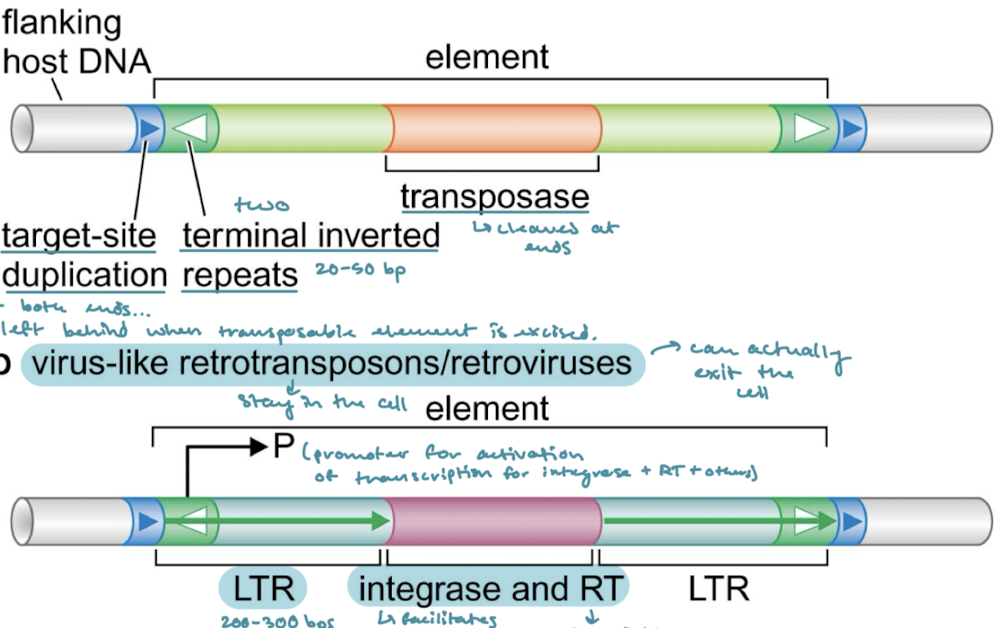
What is the difference between virus-like retrotransposons and retroviruses?
Virus-like stay in the cell, whereas retroviruses can exit the cell. Virus-like retrotransposons tend to encode for envelope genes as well so that they can interact with the membrane.
Which of the classes of transposable elements have a promoter for activation of transcription?
Virus-like retrotransposons/retroviruses and poly-A retrotransposons
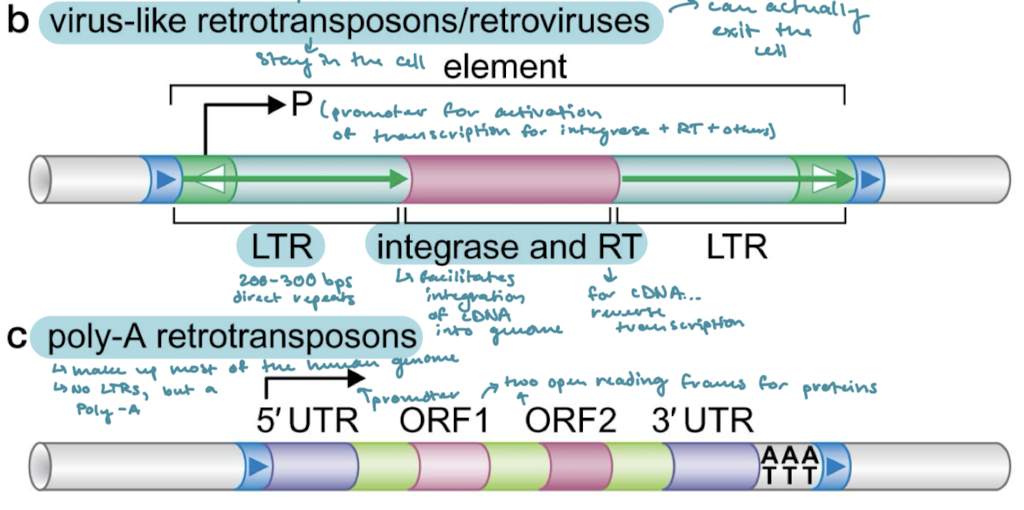
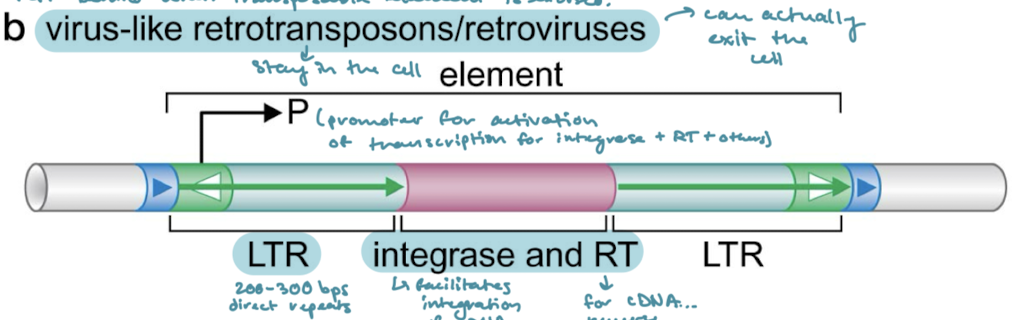
Which of the classes of transposons have long terminal repeats (LTRs)? What are LTRs?
Virus-like retrotransposons/retroviruses. 200-300 bps of direct repeats on either side of the proteins encoded for.
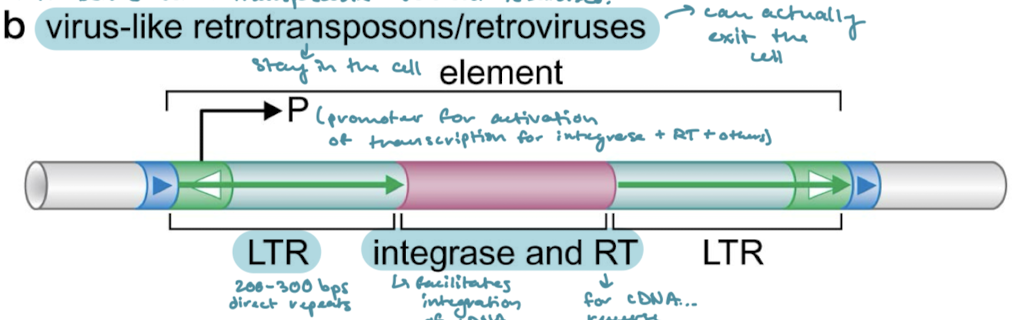
What are the proteins that are encoded in virus-like retrotransposons/retroviruses? What are their functions?
Integrase (facilitates integration of cDNA into the genome) and RT (reverse transcriptase, for cDNA synthesis)
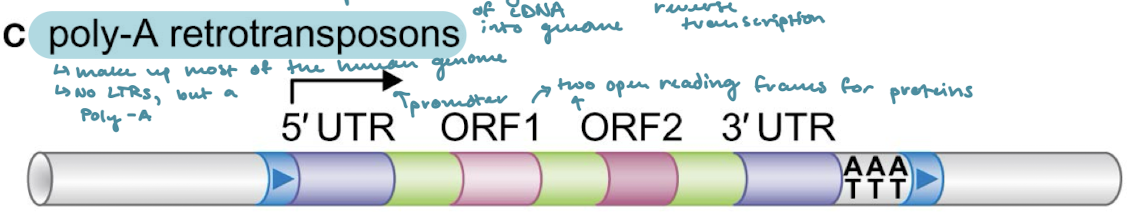
Which of the classes of transposons makes up most of the human genome?
Poly-A retrotransposons
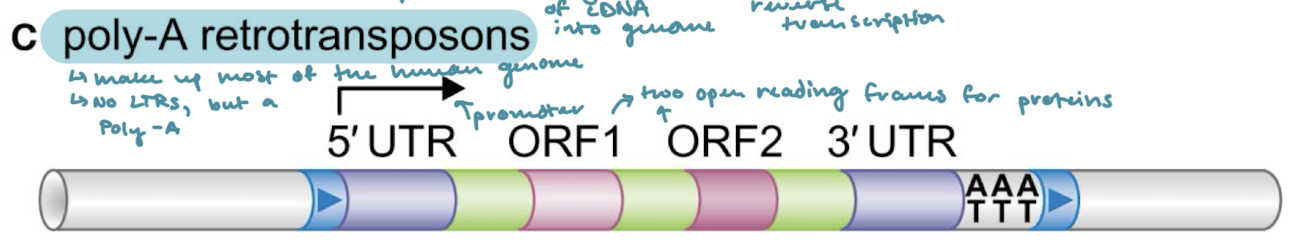
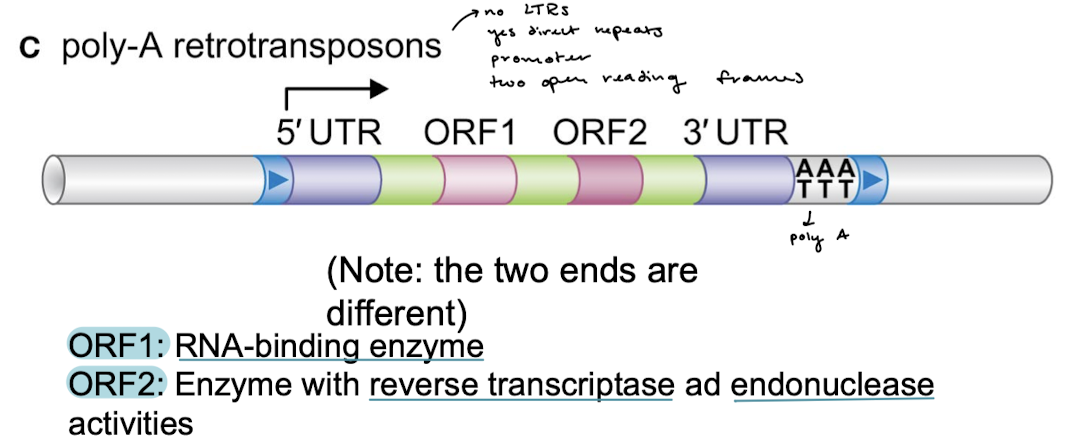
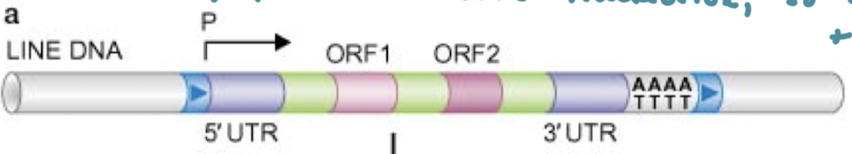
What are the three unique properties of Poly-A retrotransposons?
5’ and 3’ UTRs next to the target-site duplications (direct terminal repeats)
Two open reading frames for transcription of proteins
A poly-A on the 3’ end
Terminal inverted repeats are __________ sites
recombination
What is autonomous versus nonautonomous transposons?
Autonomous encode for functional transposase. Nonautonomous do not encode for functional transposase OR transposase is not activated. Nonautonomous transposons can receive help from an autonomous transposon’s transposase.
What are the two enzymes that Poly-A retrotransposons encode for? What do they do?
ORF1: RNA-binding enzyme
ORF2: Enzyme with reverse transcriptase and endonuclease activities.
What is cut and paste transposition?
Encode for transposase that cleaves at the inverted repeats and integrates it the element somewhere else.
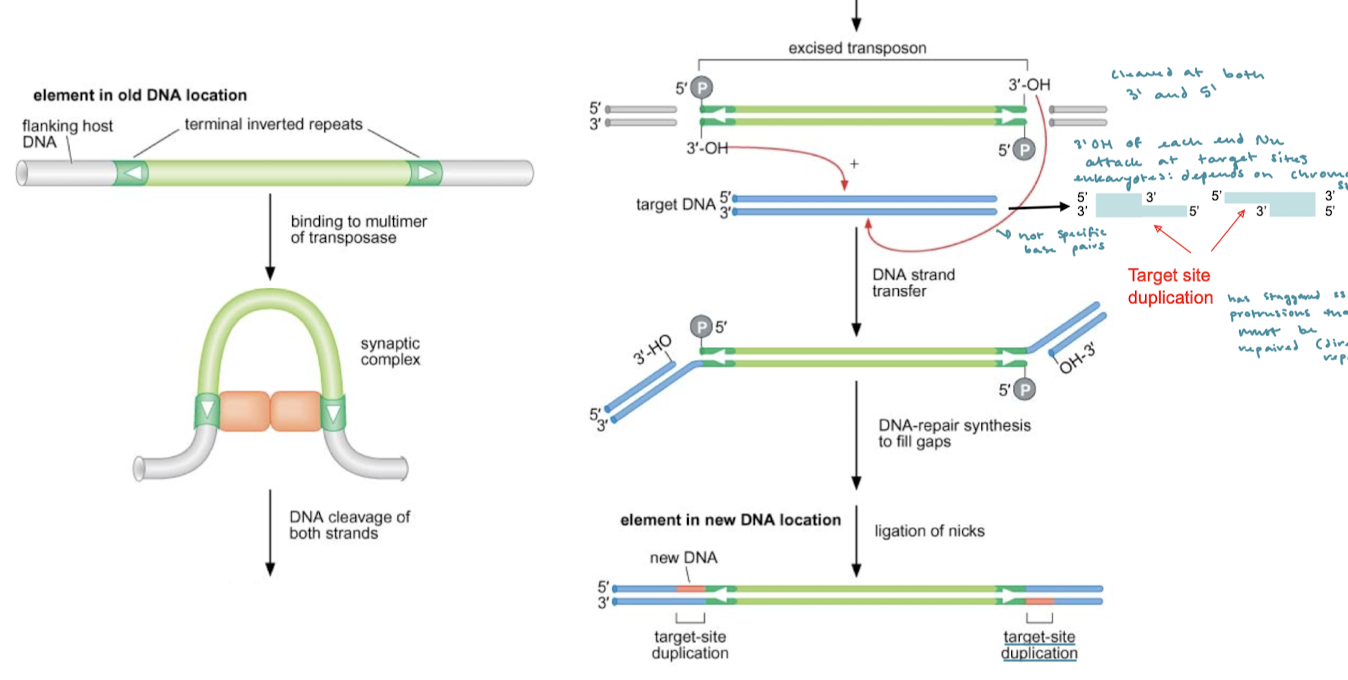
After the transposon has ben excised in cut and paste transposition, what happens next?
The 3’ OH at each end will nucleophilically attack the target sites for transfer of DNA.
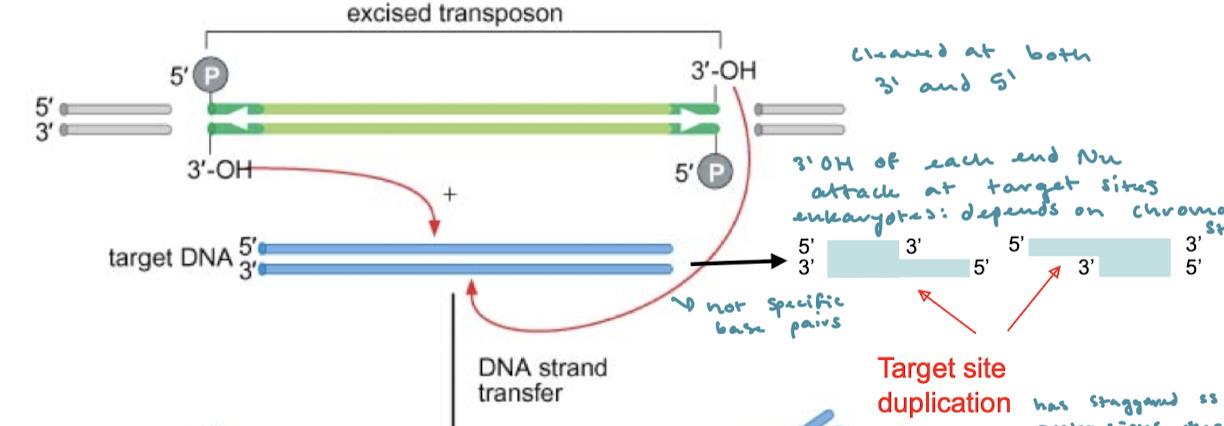
In cut-and-paste transposition, is the target DNA a specific sequence? What is another factor in eukaryotes?
The target DNA is not a specific sequence. In eukaryotes, the target site often depends on chromatin structure.
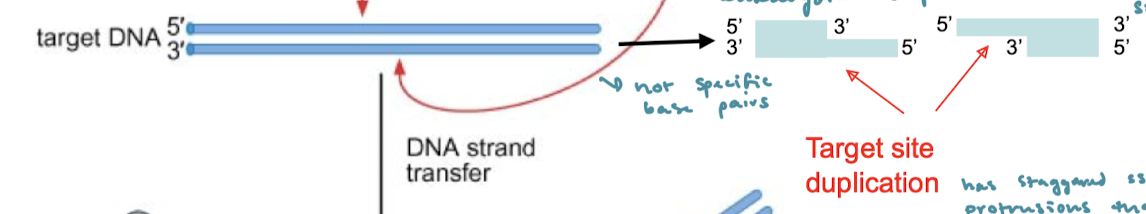
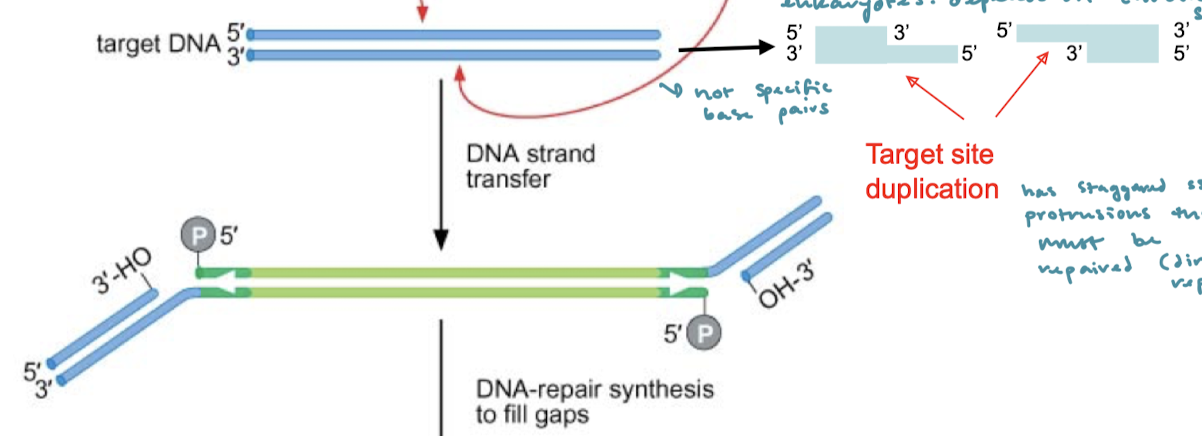
After the 3’OH nucleophilic attack on the target DNA, what are the final steps of cut-and-paste tranasposition?
There are now staggered protrusions that must be repaired. These are the target site duplications (direct repeats) that must be repaired. This is done by DNA-repair synthesis. The gaps are filled and the nicks are ligated.
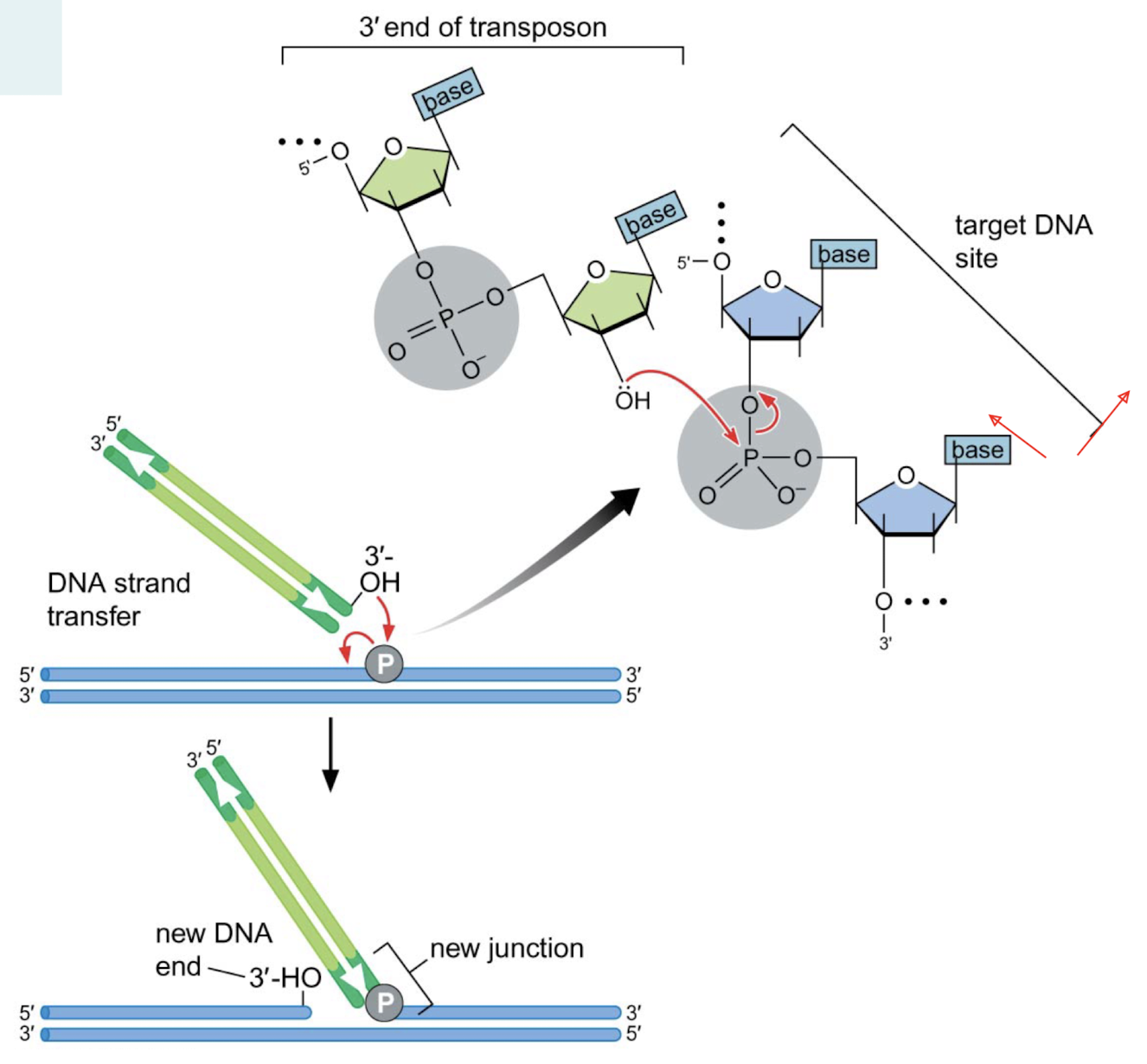
What is the chemical mechanism of DNA strand transfer?
The 3’ OH nucleophilically attacks an alpha phosphate in the phosphodiester linkage, essentially breaking that linkage at the target DNA site.
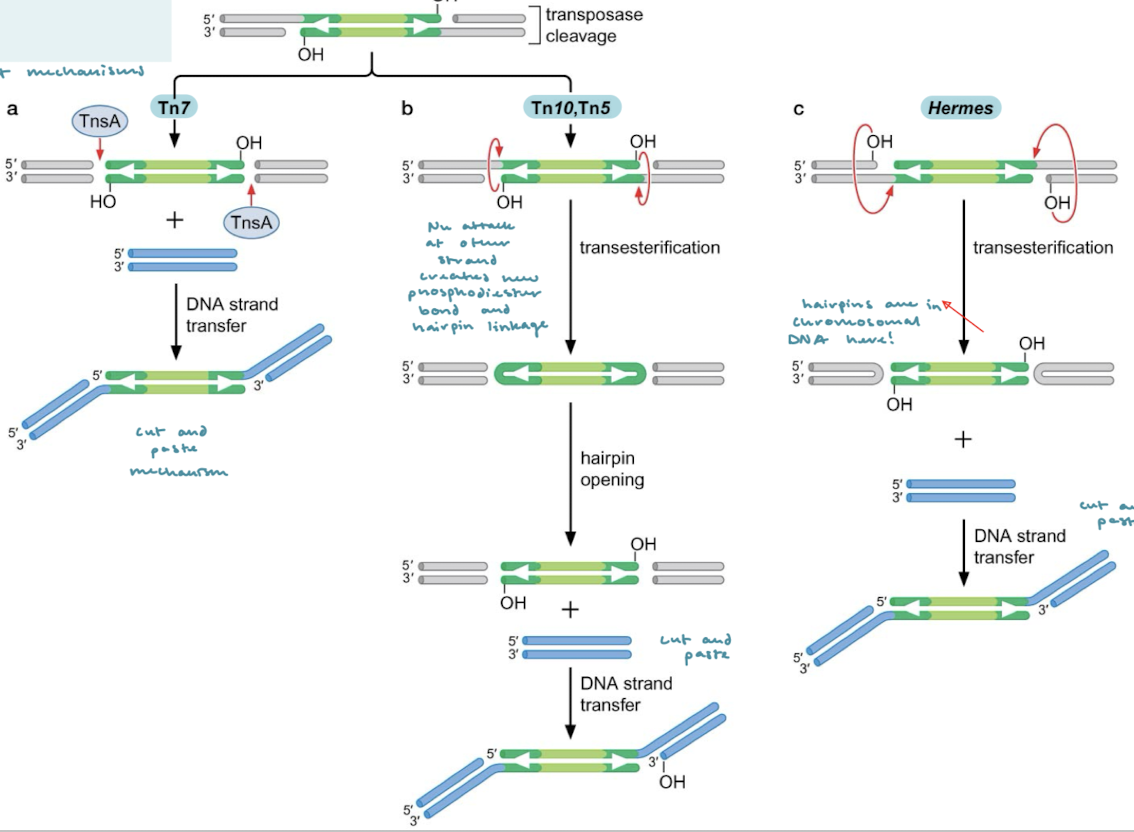
What are the three different ways of cleaving the non-transferred strand? Explain for Tn7, Tn10/Tn5, and Hermes.
Tn7: normal cut and paste mechanism using TnsA to cleave the ends.
Tn10/Tn5: Involves transesterification, a nucleophilic attack of the 3’ OH (on the transposable element) on the other strand, creating. anew phosphodiester bond and hairpin linkage. Hairpin opening is followed by the regular cut and paste mechanism.
Hermes: Transesterification, but the 3’OH is on the chromosome and not the element, making the hairpins on the chromosomal DNA. Followed by cut and paste mechanism.
Explain retroviral transposition.
DNA is transcribed into RNA. Reverse transcription results in cDNA. Integrase-catalyzed 3’ end cleavage followed by nucleophilic attack by the 3’ OH on the target DNA (catalyzed by integrase). Gap repair and ligation.
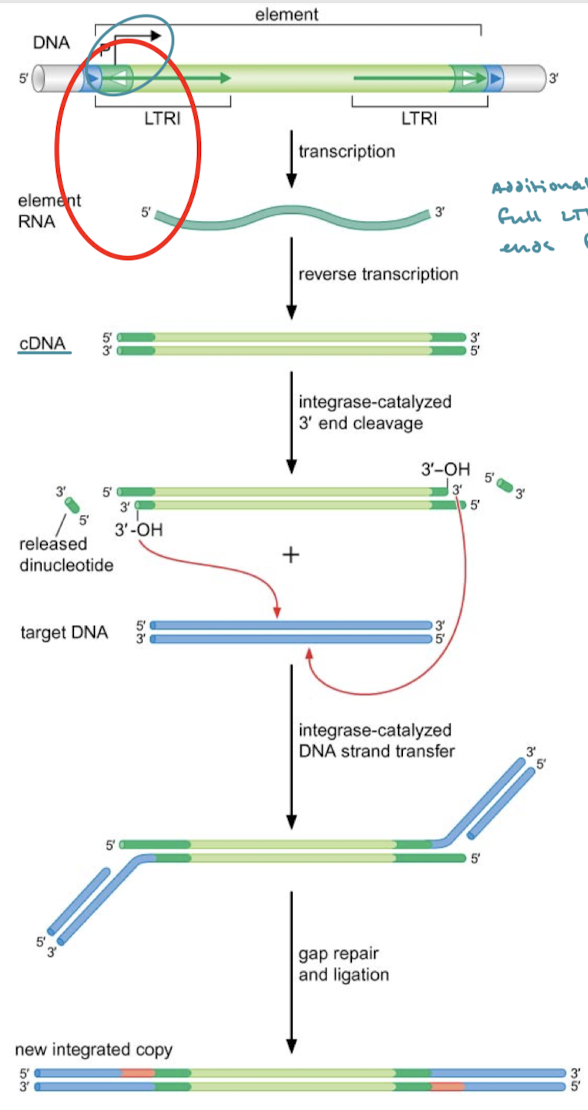
What is an example of a mechanism in retroviral transposition that ensures the full transposon is moved?
LTRs are reconstructed before transposition
Is there conservation of structure and function in transposases and integrases?
Yes
What is the DDE transposase superfamily?
A large group of enzymes, categorized by their conserved “DDE” amino acid motif, responsible for catalyzing the movement of DNA transposons within a genome.
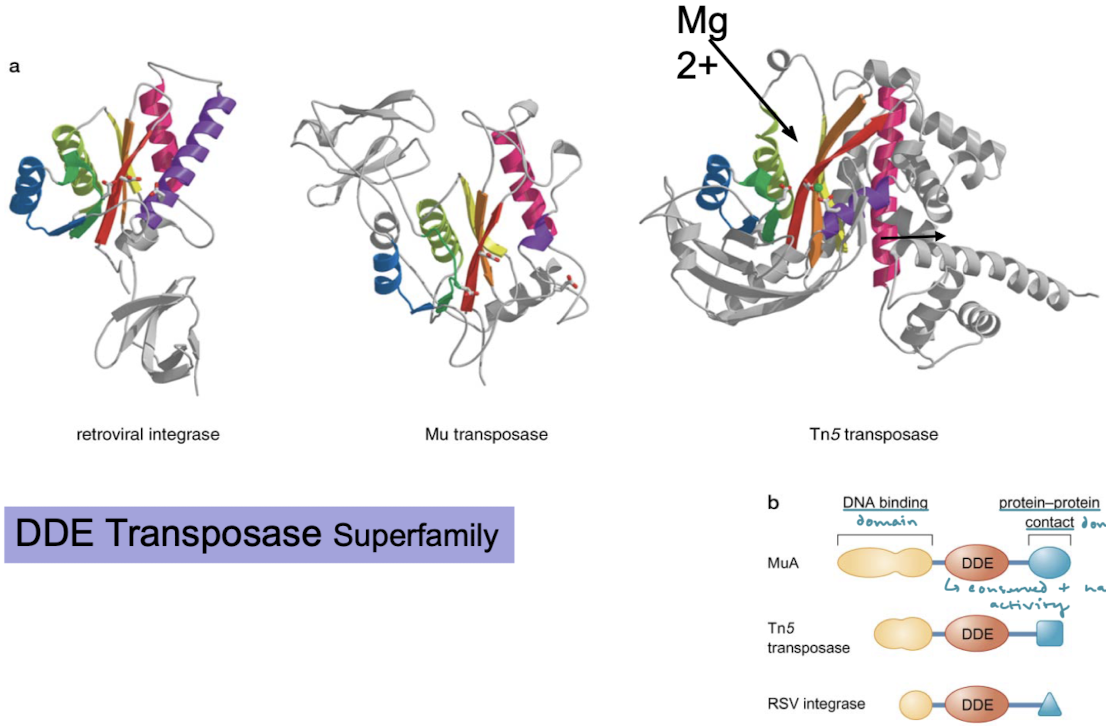
What is the well-conserved domain of transposases? What are the other domains present?
The catalytic domain (DDE motif) is well conserved and has integrase activity. There are also DNA binding domains and protein-protein contact domains for functioning as a complex.
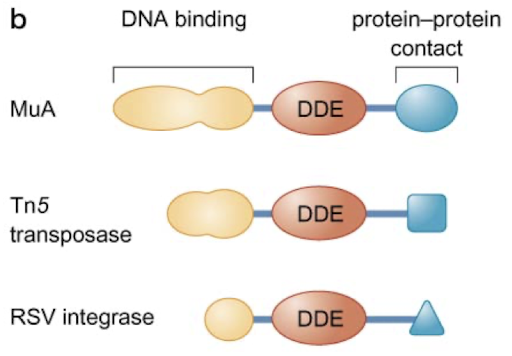
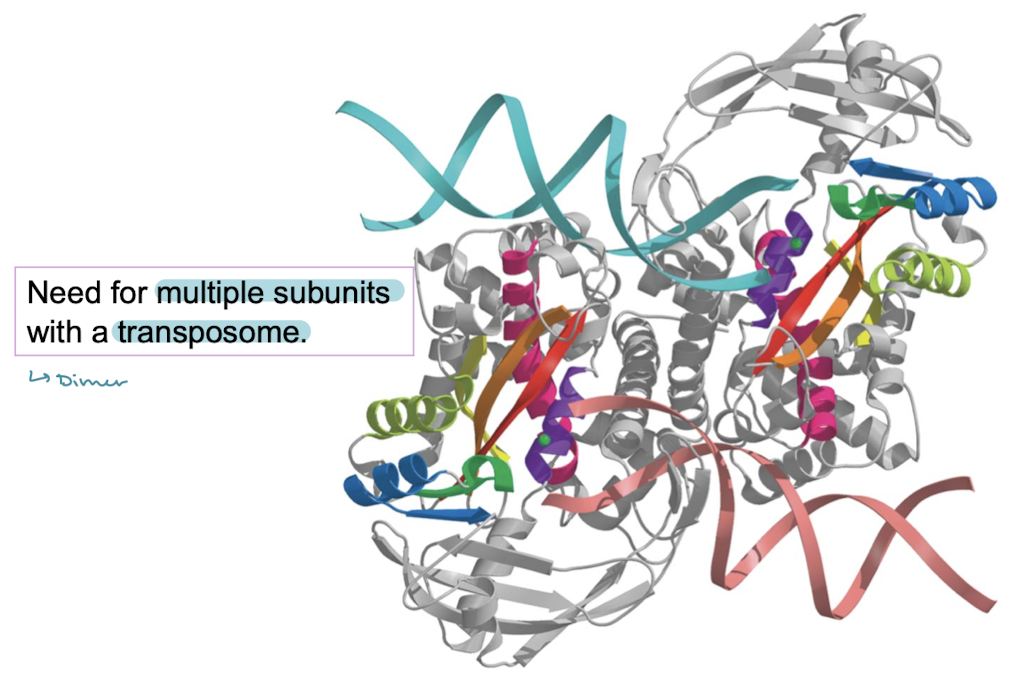
Tn5 transposase has a need for multiple ______ with a _______.
subunits (dimer); transposome
Poly-retrotransposons use reverse transcription. Do they use the cut and paste mechanism? What is another name for them?
No. They are also called human LINE elements (long interspersed elements).
What protein is involved in RT by poly-A retrotransposons? What activities does this protein have?
ORF2 protein: endonuclease and RT activity

The phrase “target-site primed reverse transcription” is associated with what transposable element?
RT by Poly-A retrotransposons
What is the difference between the promoters of virus-like retrotransposons/retroviruses and Poly-A retrotransposons?
The promoter of poly-A retrotransposons is located right at the first nucleotide, so mRNA is a full and intact copy of the element, including the Poly A tail. This is not the case in virus-like and retroviruses.
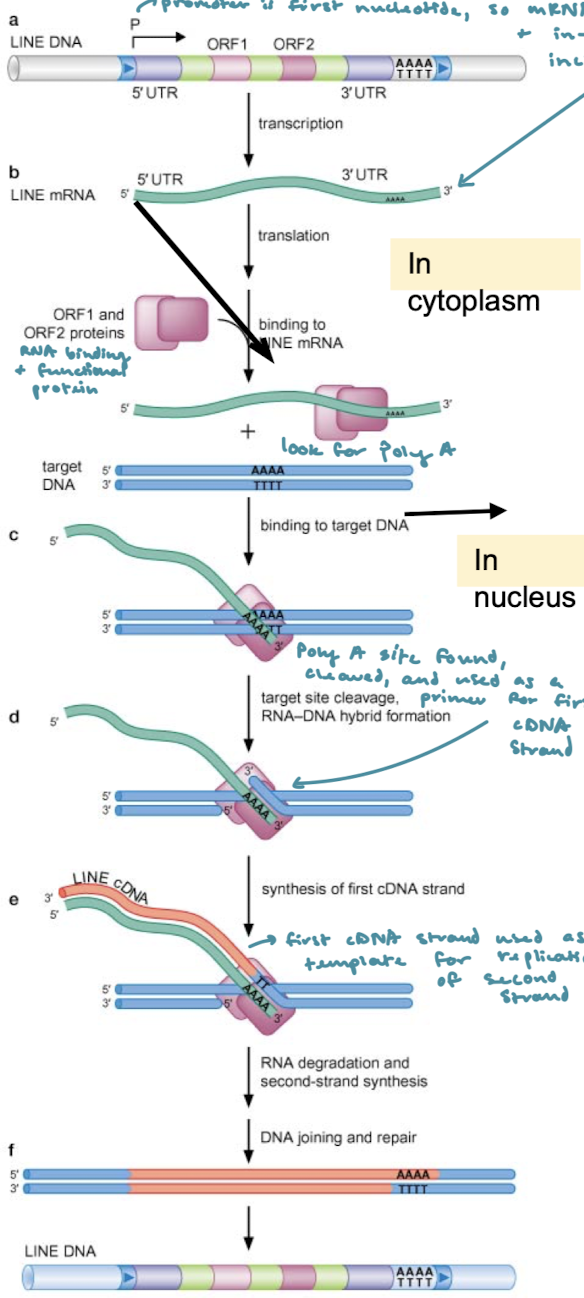
In RT of poly-A retrotransposons, what happens after LINE mRNA has been transcribed?
ORF1 (RNA binding protein) and ORF2 (endonuclease and RT activity) proteins bind to the mRNA and search for the Poly A portion (cytoplasm). The poly-A site is found, cleaved, and used as a primer for the first cDNA strand (nucleus). The first cDNA strand is used as a template for replication of the second strand and the RNA is degraded. DNA joining and repair.
What are the two theories behind why transposable elements are so abundant when they are responsible for/cause mutations?
Selfish DNA hypothesis: a mechanism to survive in future generations (parasitic)
Could the genome be benefiting? Yes! Transposons can change the activation of gene promoters (or other regulations) that are beneficial. Genes can also contain transposable elements and obtain a new and favorable function that can be passed on through a lineage. Also beneficial for the variation of antibodies in the immune system.
V(D)J recombination occurs in what region of the antibody? Why is this process important?
The variable region is where particular antigens are recognized. Recombination is important for the specificity of antigen binding.
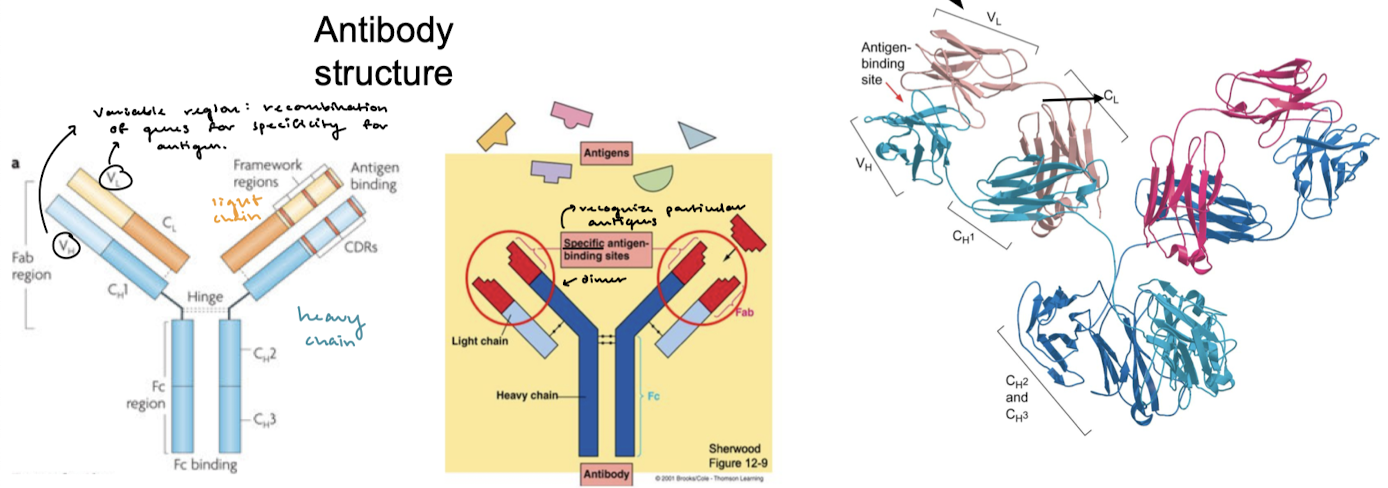
Which of the chains contains the diversity segments?
The heavy chain contains diversity segments. V, J, and C segments are common to both light and heavy chains.
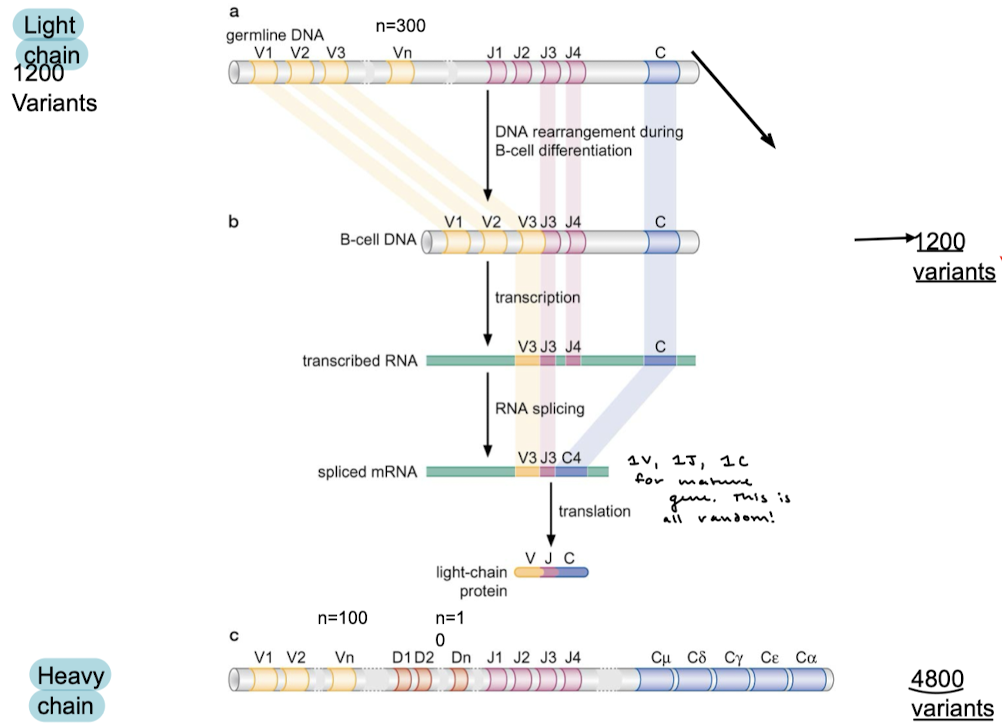
What proteins mediate gene rearrangements in V(D)J recombination?
RAG proteins
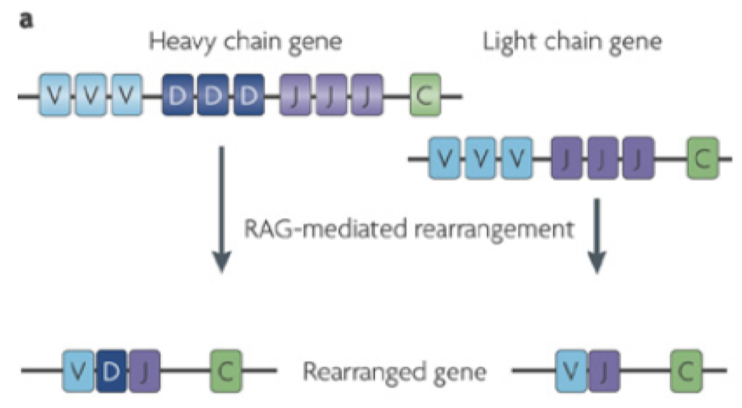
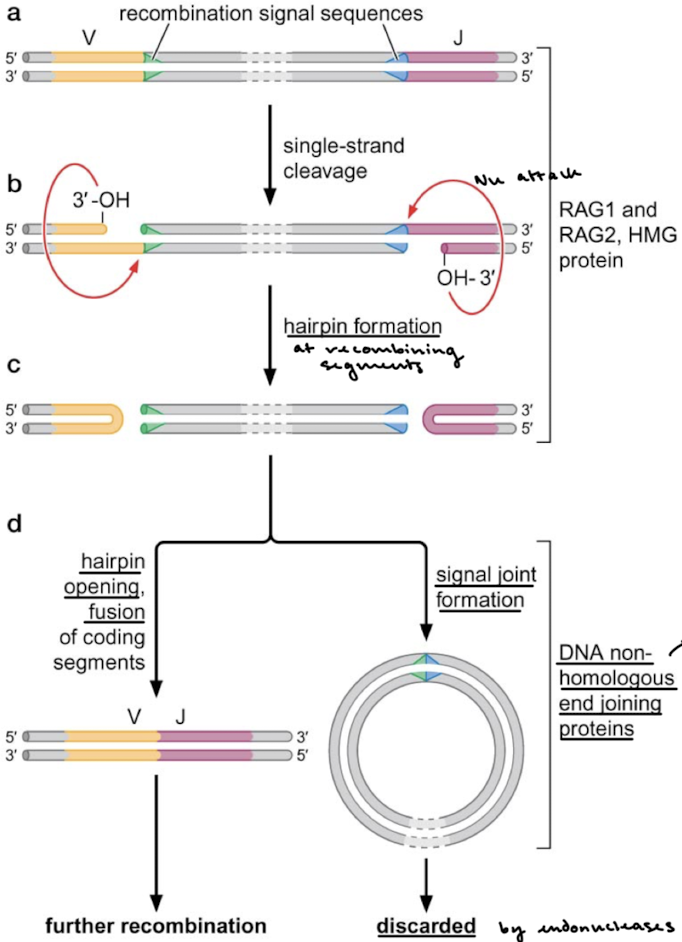
What is required for V(D)J recombination by RAG1/RAG2? What is this rule called?
Recombination signal sequences. This rule is called the 12/23 rule, where recombination always occurs between a 12-bp spacer recombination signal sequence and a 23-bp spacer recombination signal sequence.
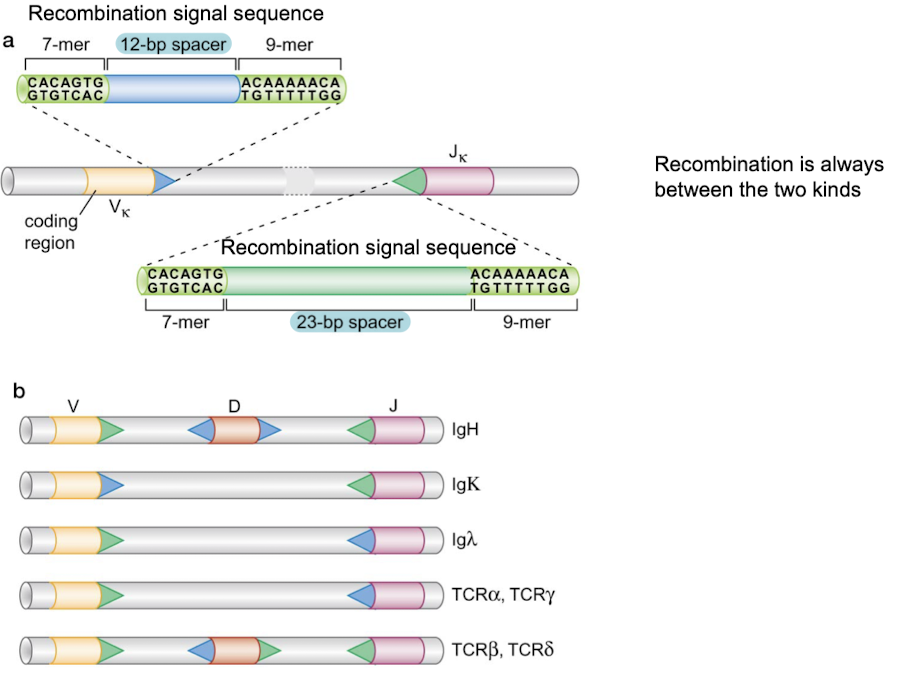
RAG1 and RAG2 recombinases function much like what enzyme?
transposase
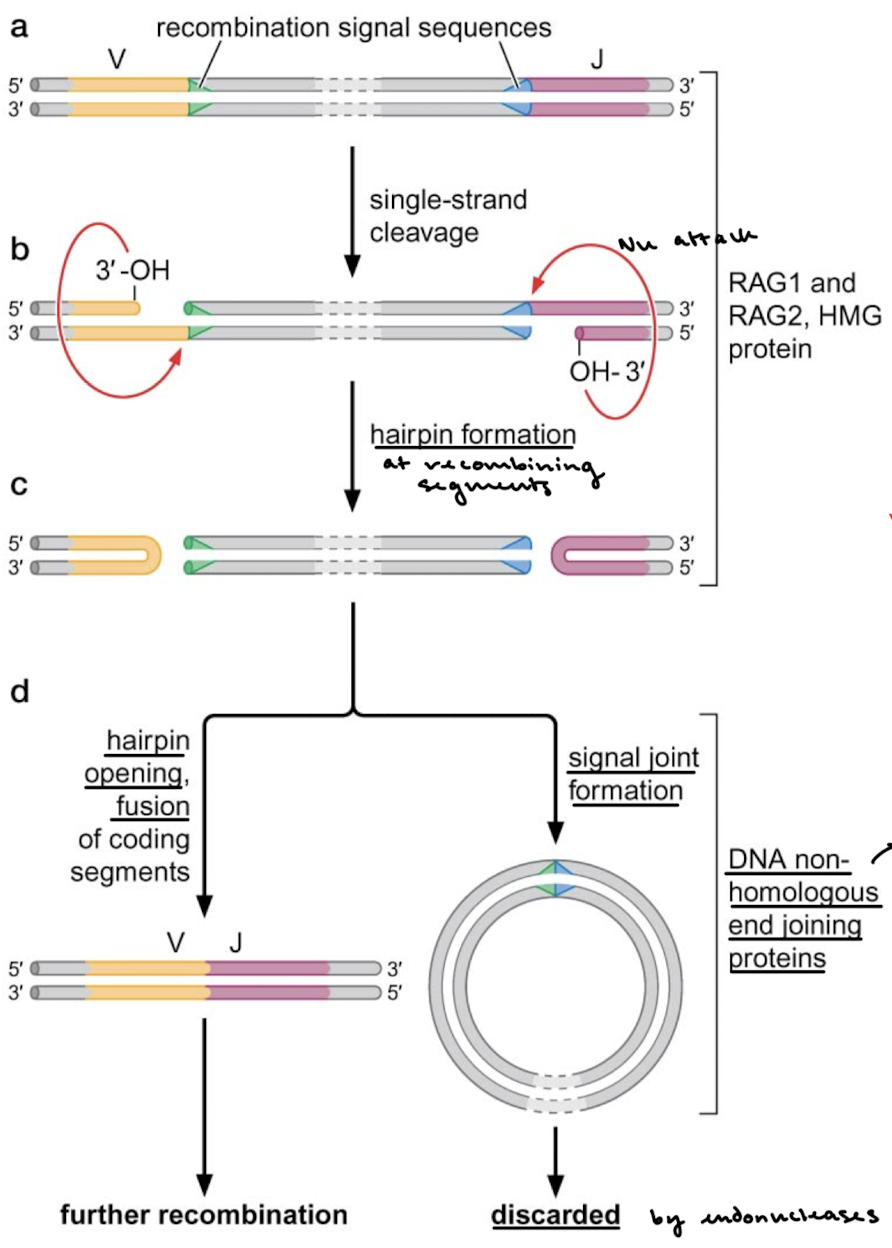
Explain the mechanism of RAG1 and RAG2-mediated recombination. What mechanism is responsible for joining the segments and the signal joints?
3’OH (on the recombining segments) nucleophilically attacks the other strand, forming a hairpin at the recombining segments. Hairpin opening and fusion. of coding segments occurs and recombination continues. The signal sequences (and all of the DNA between them) form a signal joint and are discarded by endonucleases. NHEJ is responsible, which creates more mutations and adds to the variability of the variable ends.