Cell Bio Ch 4
1/123
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
124 Terms
enzymes function
catalyze covalent bond formation or breakage
structural proteins function
provide mechanical support to cells and tissues
transport proteins function
carry small molecules/ions
motor proteins function
generate movement in cells and tissues
storage proteins function
store amino acids or ions
signal proteins function
carry extracellular signals from cell to cell
receptor proteins function
detect signals and transmit them to cell’s response
transcription regulators function
machinery bind DNA to switch genes on or off
special purpose proteins function
highly variable
what are proteins made of
amino acids
order of amino acid sequence unique or diff for each protein
unique
linear sequence of amino acid considered
primary structure
the r group of amino acid determines
identity/chemical behavior
How many amino acids in polypeptide chain
20
how do polymers grow
the addition of a subunit onto one end of the polymer chain via a condensation reaction, where a molecule of water is lost for each subunit added
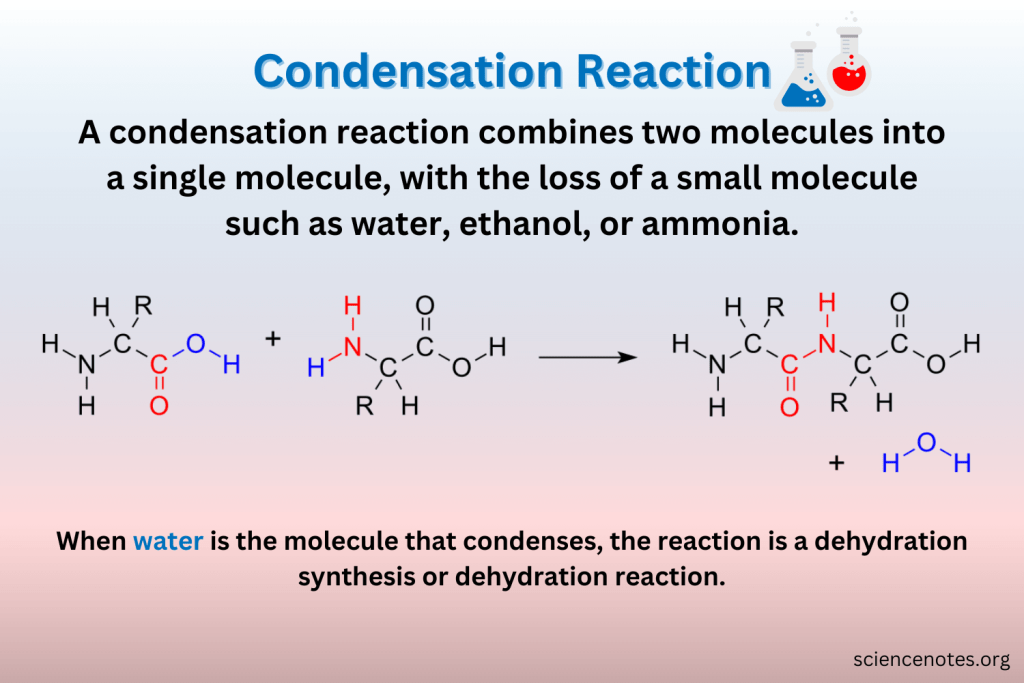
what holds amino acid in protein together
peptide bonds between carboxyl and amino group of adjacent amino acid
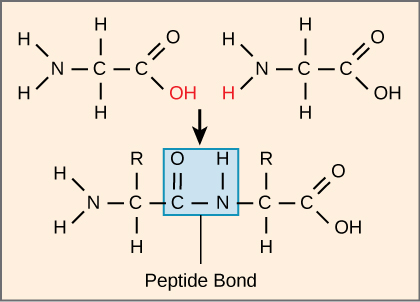
proteins are also known as polypeptides because
they made of peptide bonds
protein is made of
amino acids linked in polypeptide chain
directionality of protein synthesis is known as
N to C synthesis
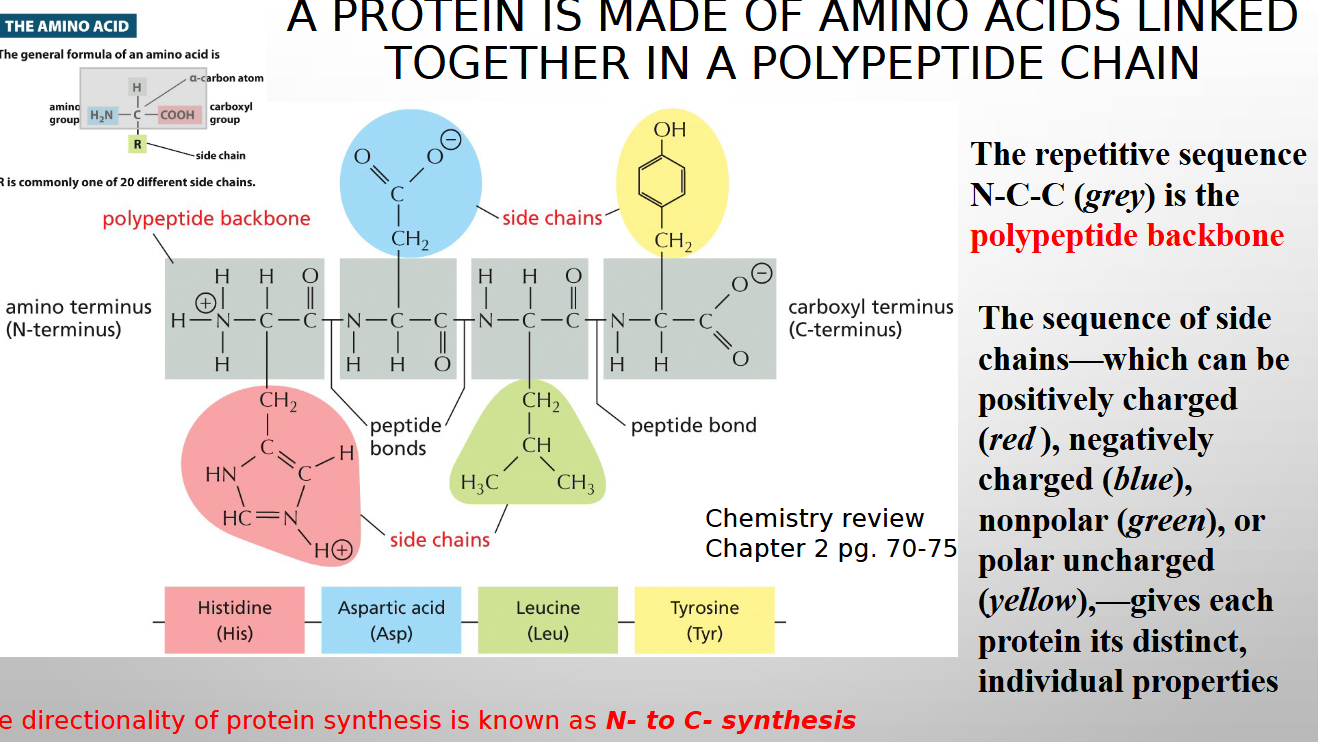
20 common amino acids
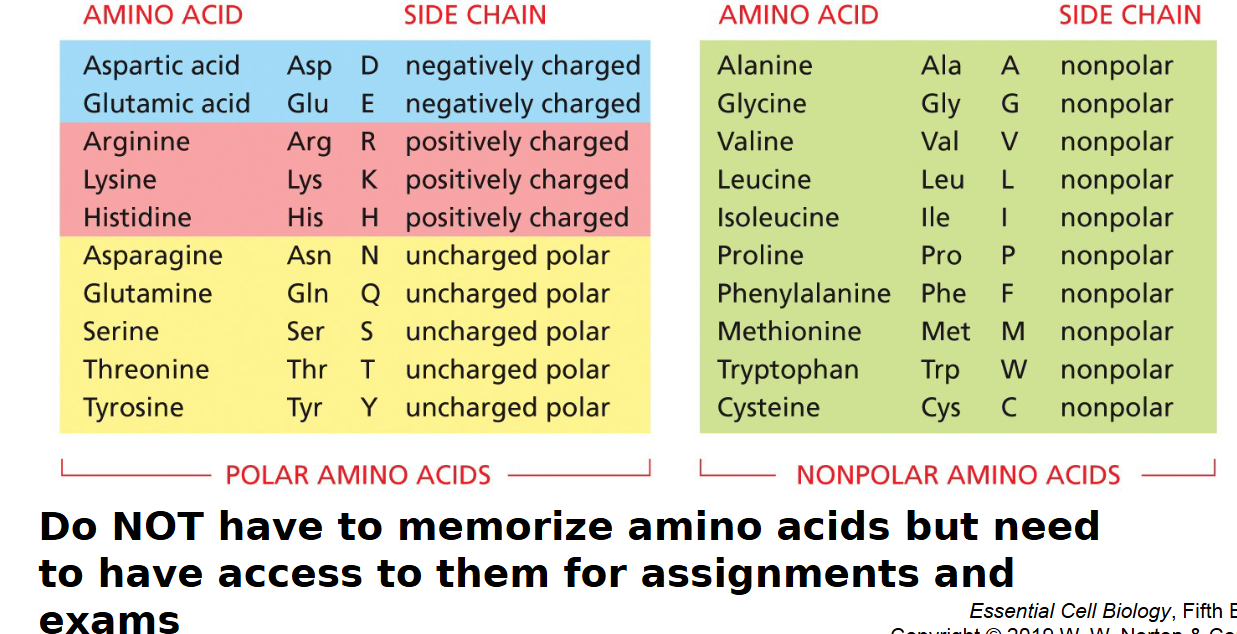
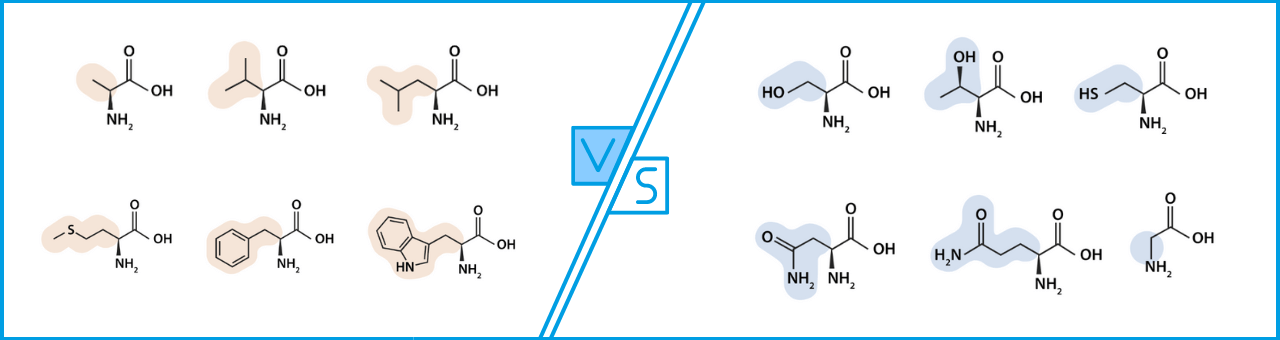
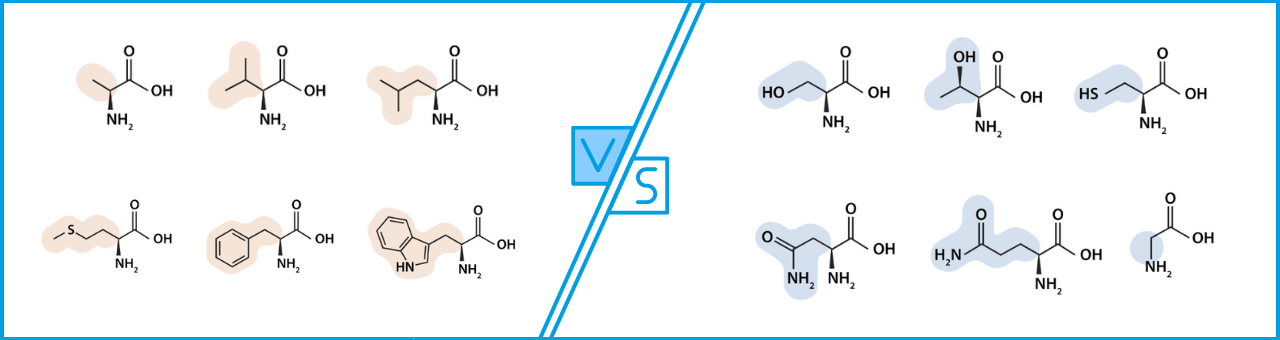
polar and charged amino acids vs nonpolar amino acids
hydrophilic vs hydrophobic
polar side chains of polypeptide bond
partial charges can form hydrogen bonds
electrically charged side chains
charged side chains can form ionic and polar bonds
three types of non covalent bonds that help proteins fold (HEV)
hydrogen bonds, electrostatic attractions, vanderwal attractions
Hydrophobic forces help proteins
fold into compact conformations
how is a hydrophobic core formed
Hydrophobic amino acid side chains cluster together in the
protein's interior, away from the surrounding water, forming a hydrophobic core
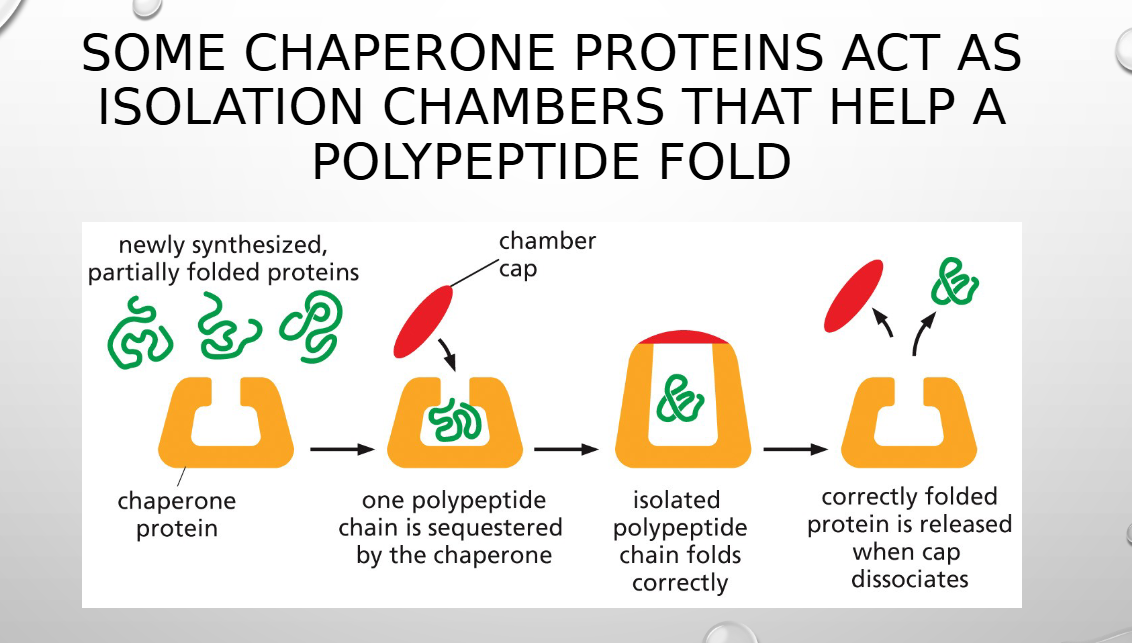
chaperone proteins do what
assist in folding the polypeptide chain (folding decreases conformation energy)
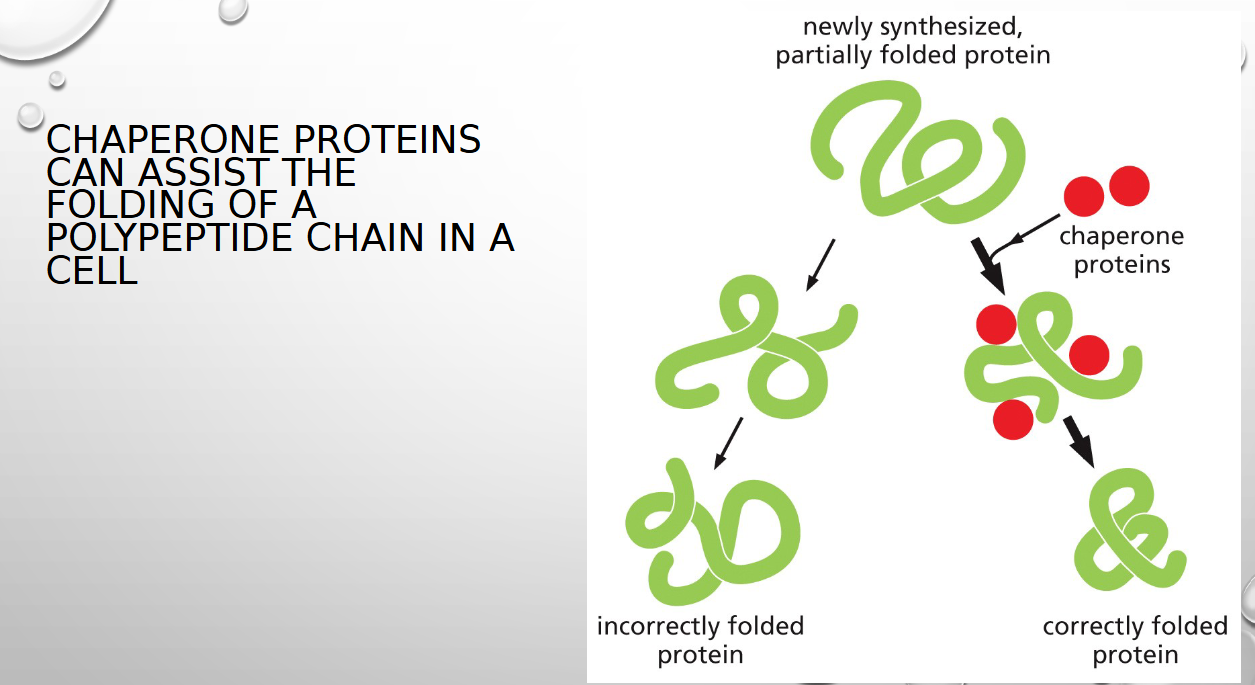
some chaperone proteins help with folding by forming
isolation chambers
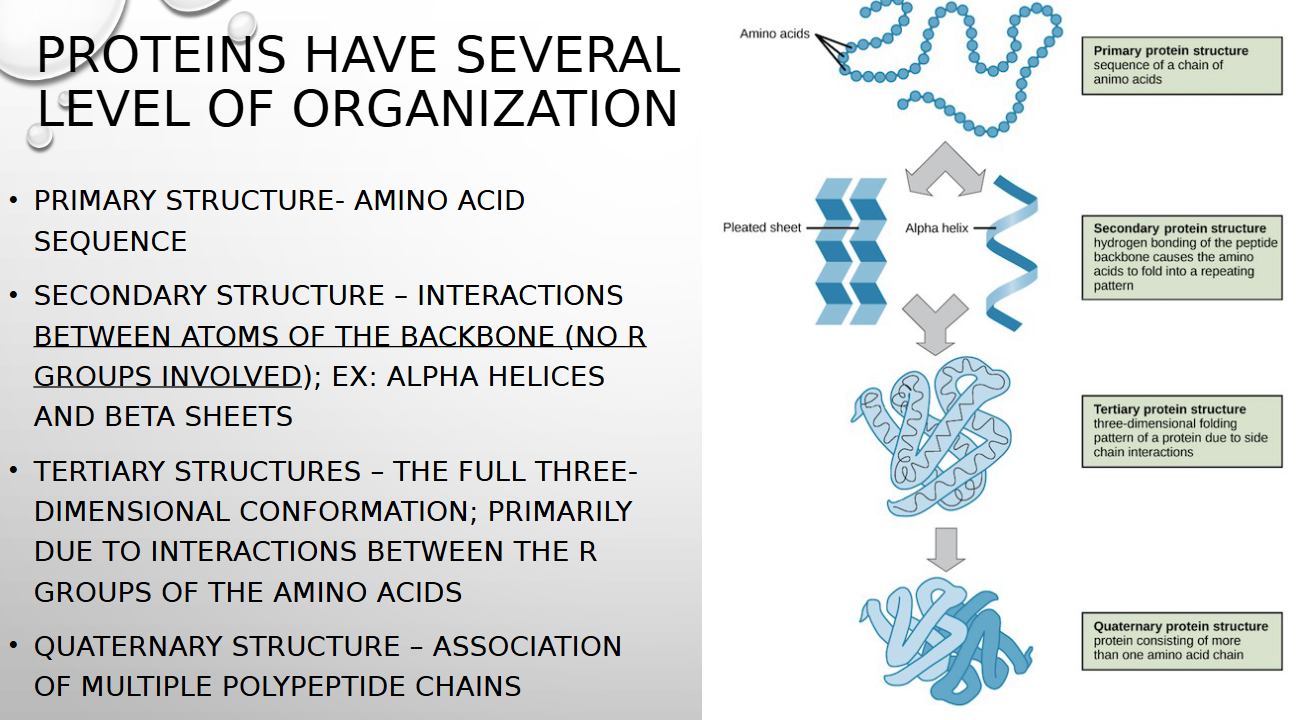
primary vs secondary protein structure
amino acid sequence vs interactions between atoms of backbone (NO R GROUP INVOLVED)
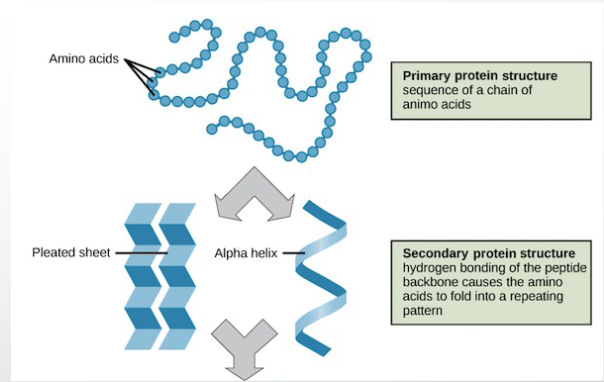
tertiary structure
full 3d conformation primarily due to interactions between r groups of amino acids
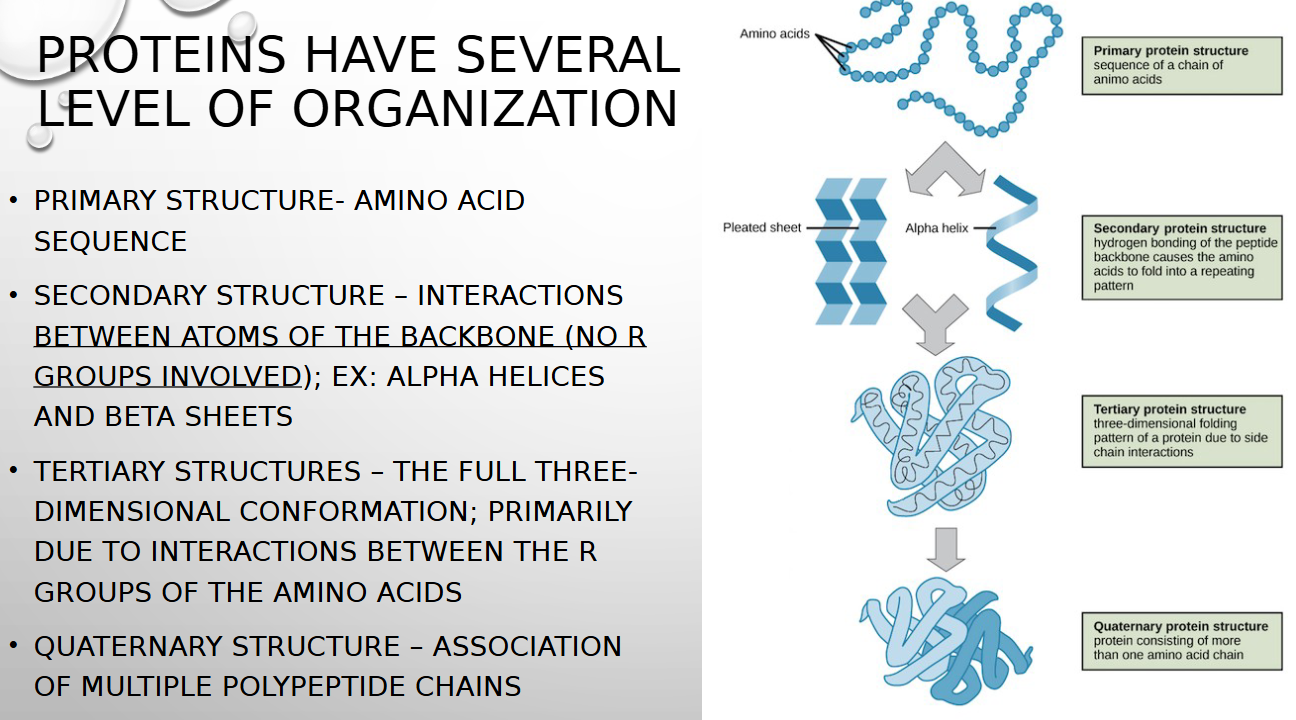
quaternary structure
ASSOCIATION OF MULTIPLE POLYPEPTIDE CHAINS
what does it mean to say R group is not involved for secondary protein structure
hyd bonds involve ONLY the atoms of the backbone
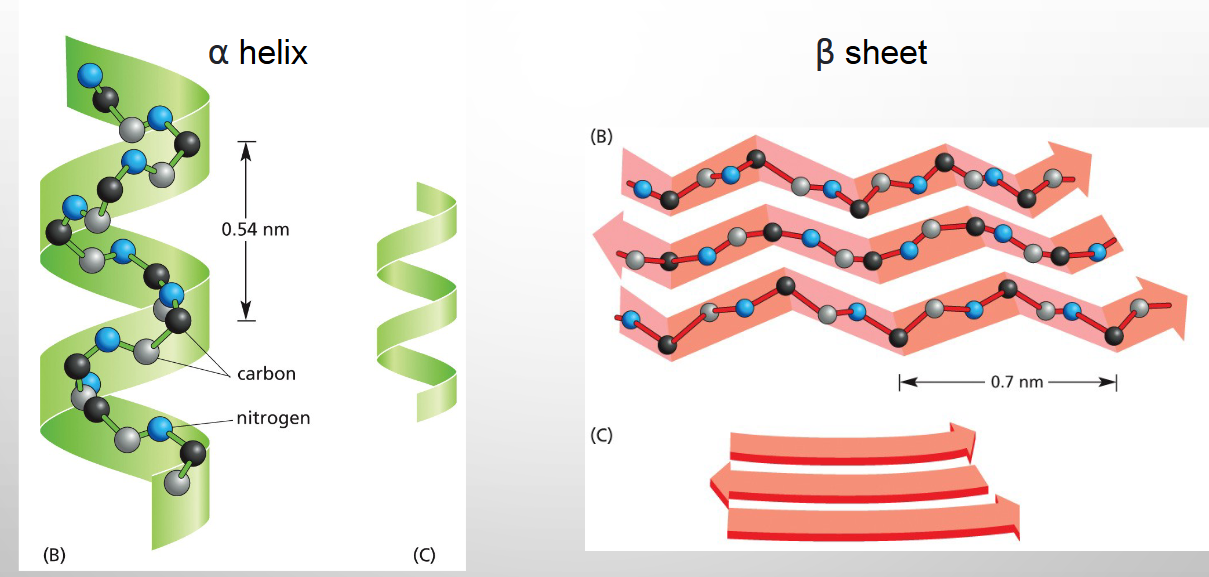
N–H of every peptide bond is hydrogen-bonded to the C=O of a neighbouring peptide bond located _____ amino acids
away in the same chain.
FOUR
α-HELICES FORM CYLINDERS THAT CAN CROSS
LIPID BILAYERS
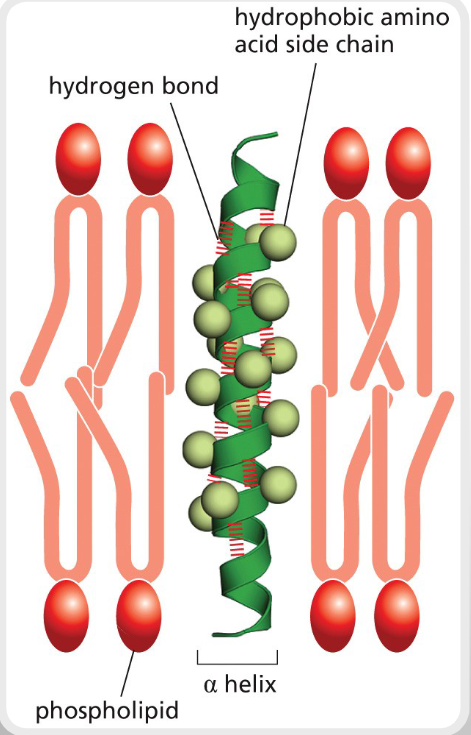
PROTEINS EMBEDDED IN THE CELL MEMBRANE OFTEN CONTAIN
SHORT ΑLPHA HELICES STRUCTURES (I.E. RECEPTORS, TRANSMEMBRANE PROTEINS, AND TRANSPORT PROTEINS)
THE R SIDE CHAINS THAT STICK OUT THE POLYPEPTIDE ARE (polar or nonpolar)
NON-POLAR (HYDROPHOBIC)
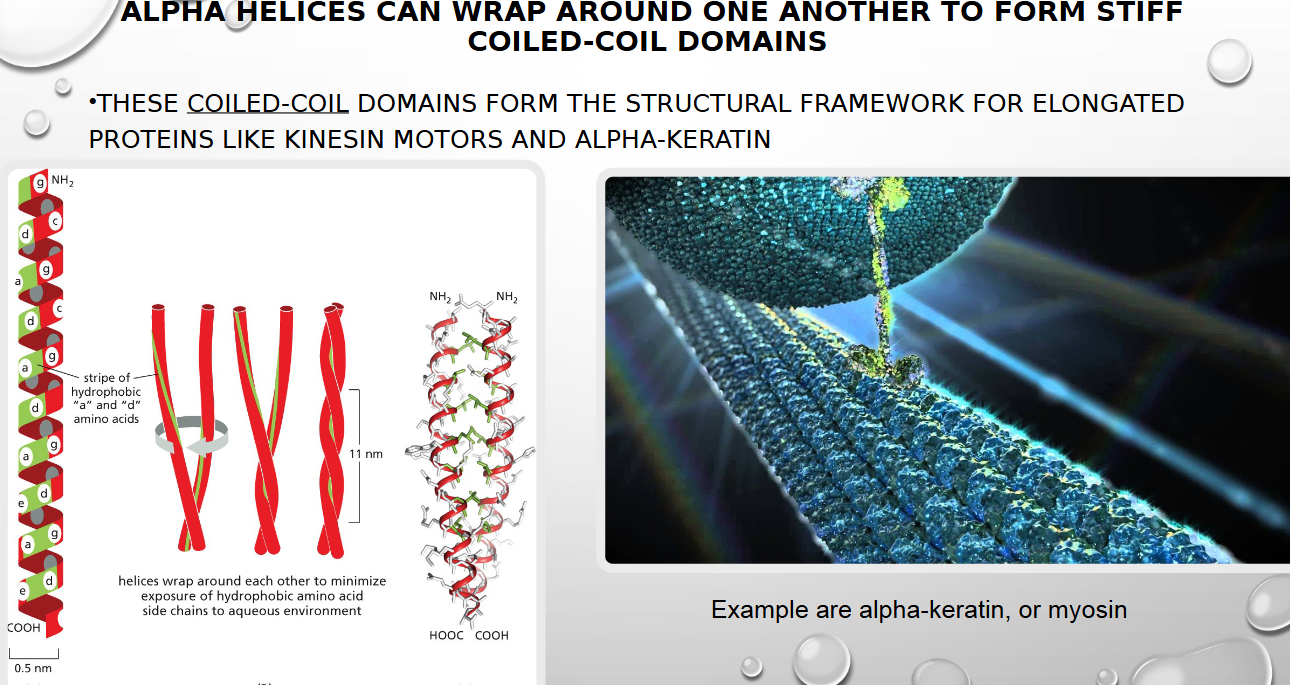
ALPHA HELICES CAN WRAP AROUND ONE ANOTHER TO FORM STIFF
COILED-COIL DOMAINS
the stiff coiled coil domains alpha helices form often make what type of proteins
ELONGATED ONES like keratin
Hydrogen bonds between N-H and to C=O happen between
BACKBONE atoms of adjacent strands (no r group involved)
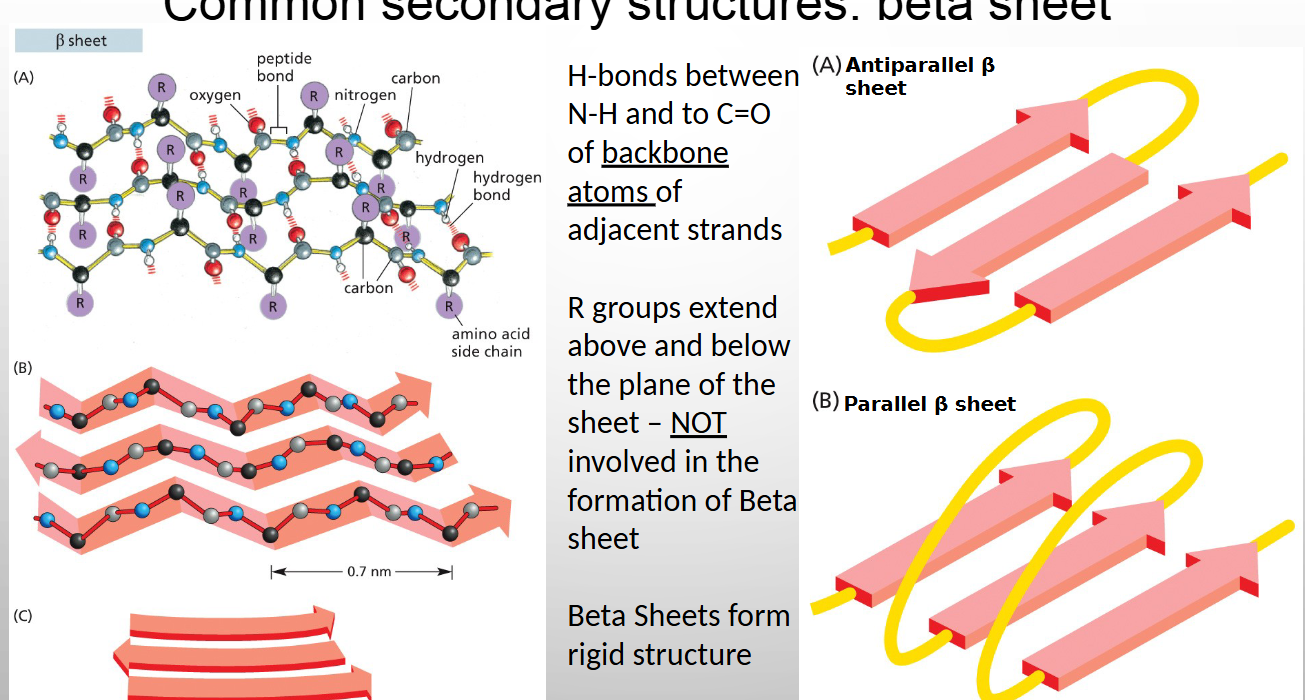
separate regions of proteins are called
domains
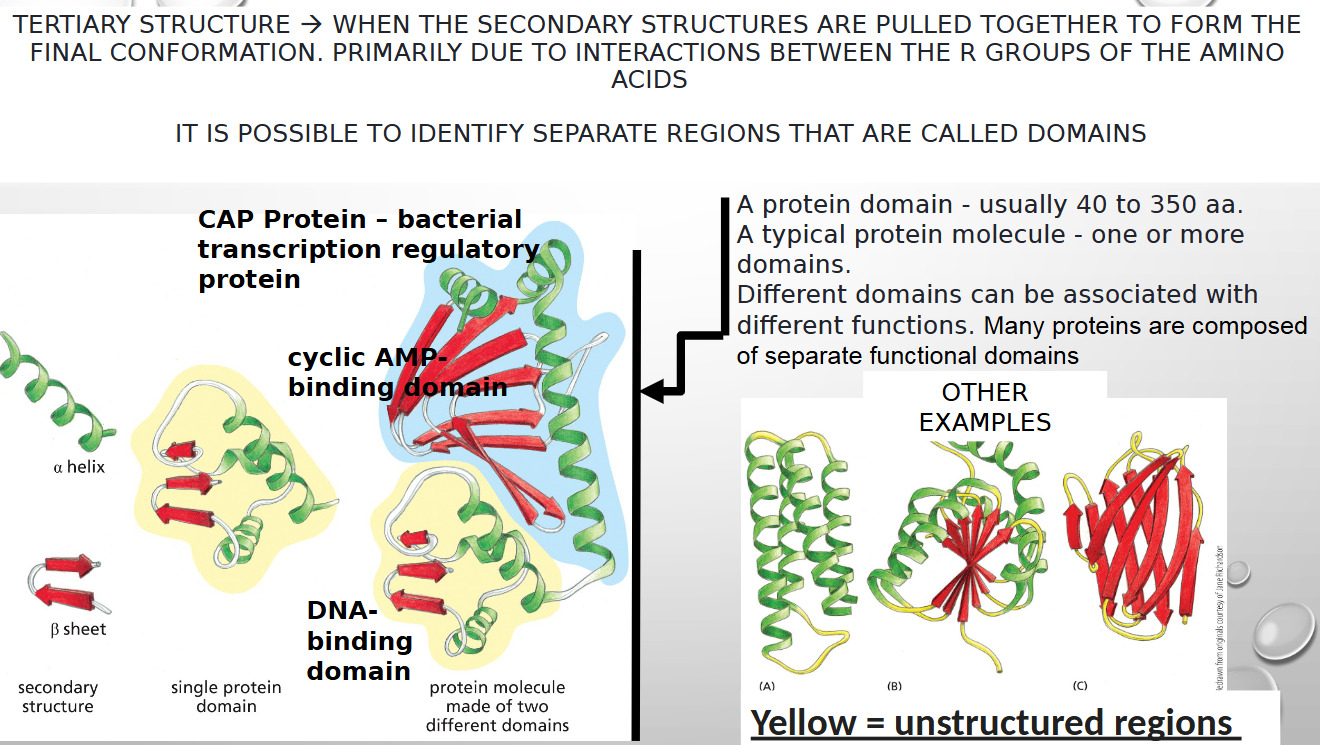
WHY ARE THE ALPHA HELIX AND BETA SHEET SO COMMON IN PROTEIN STRUCTURE?
their structures are stabilized by stable, regular hydrogen bonds between the backbone atoms of the polypeptide chain (NOT DEPENDENT UPON A PARTICULAR SIDE CHAIN)
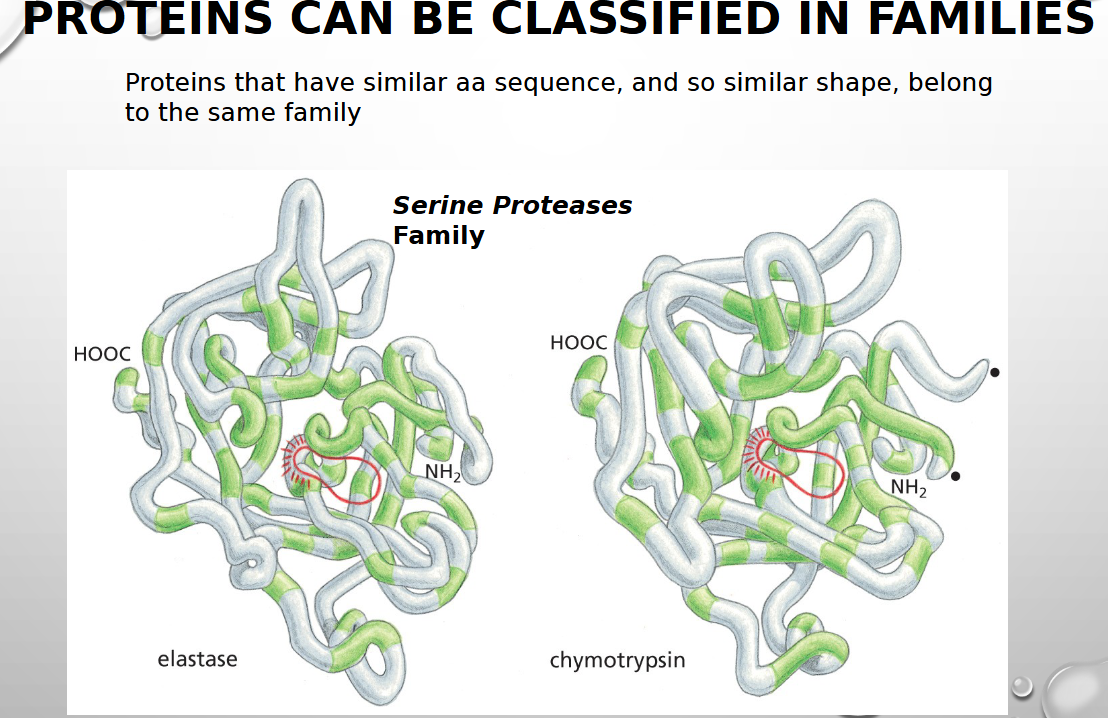
Proteins that have similar aa sequence, and so similar shape, belong to the same
family
EACH DIFFERENT POLYPEPTIDE CHAIN IS CALLED A
SUBUNIT
REGIONS OF PROTIENS INVOLVED IN MOLECULAR INTERACTION ARE CALLED
BINDING SITES
HEMOGLOBIN
AN OXYGEN- CARRYING PROTEIN ABUNDANT IN RED BLOOD CELLS,
HEMOGLOBIN CONTAINS TWO COPIES OF WHAT GLOBINS
TWO α-GLOBIN (GREEN) AND TWO COPIES OF β-GLOBIN (BLUE)
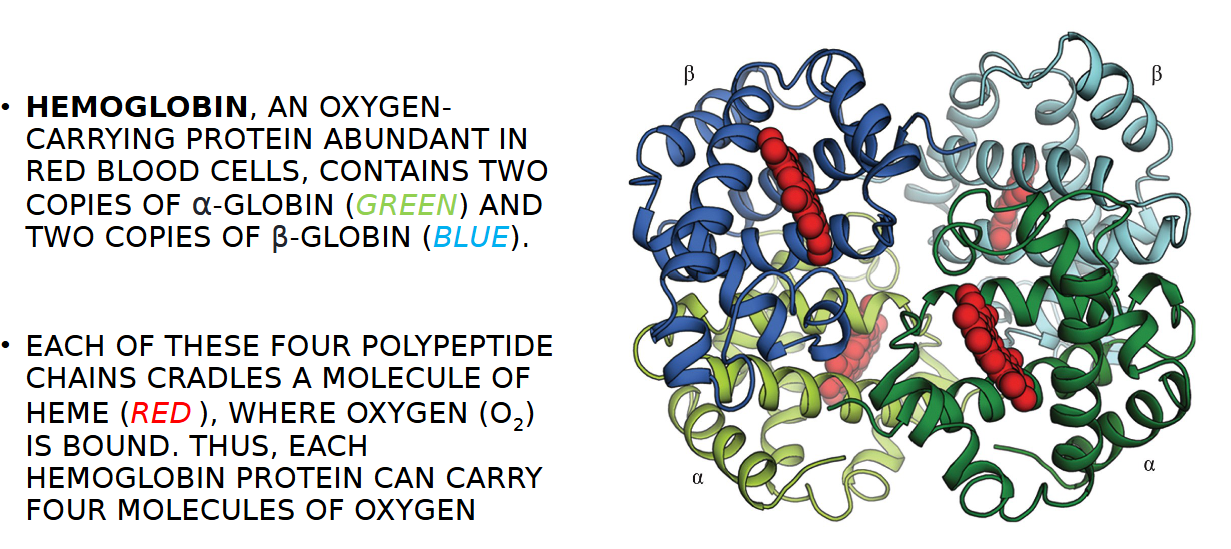
HOW MANY MOLECULES OF OXYGEN PER HEMOGLOBIN PROETIN
EACH OF THESE FOUR POLYPEPTIDE CHAINS CRADLES A MOLECULE OF HEME (RED ), WHERE OXYGEN (O2) IS BOUND. THUS, EACH HEMOGLOBIN PROTEIN CAN CARRY FOUR MOLECULES OF OXYGEN
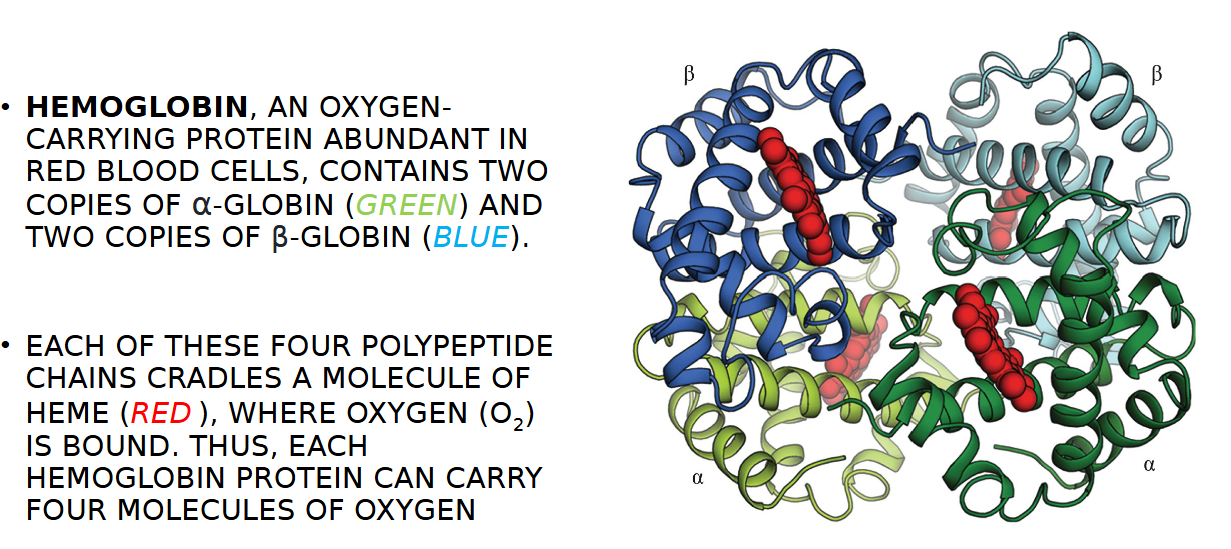
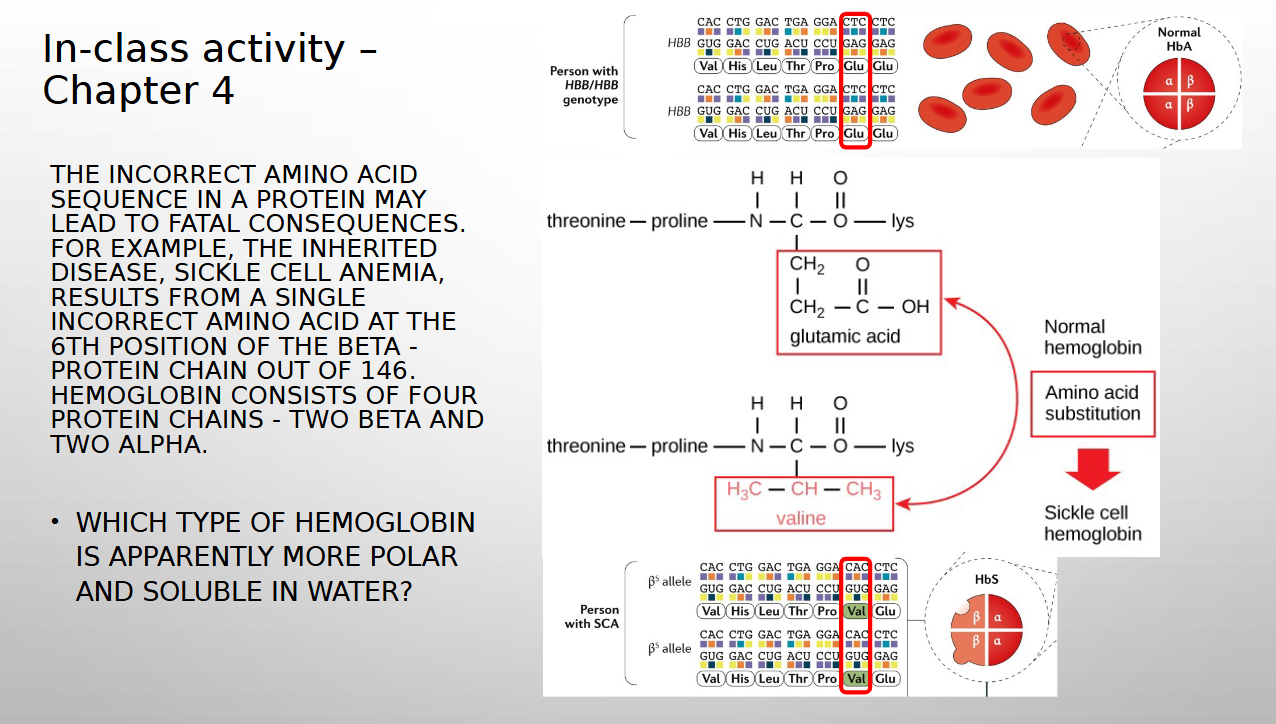
try to answer
Normal hemoglobin (HbA) is more polar and soluble in water than the sickle cell hemoglobin (HbS) because HbA has glutamic acid which loves water and HbS has valine which hates water
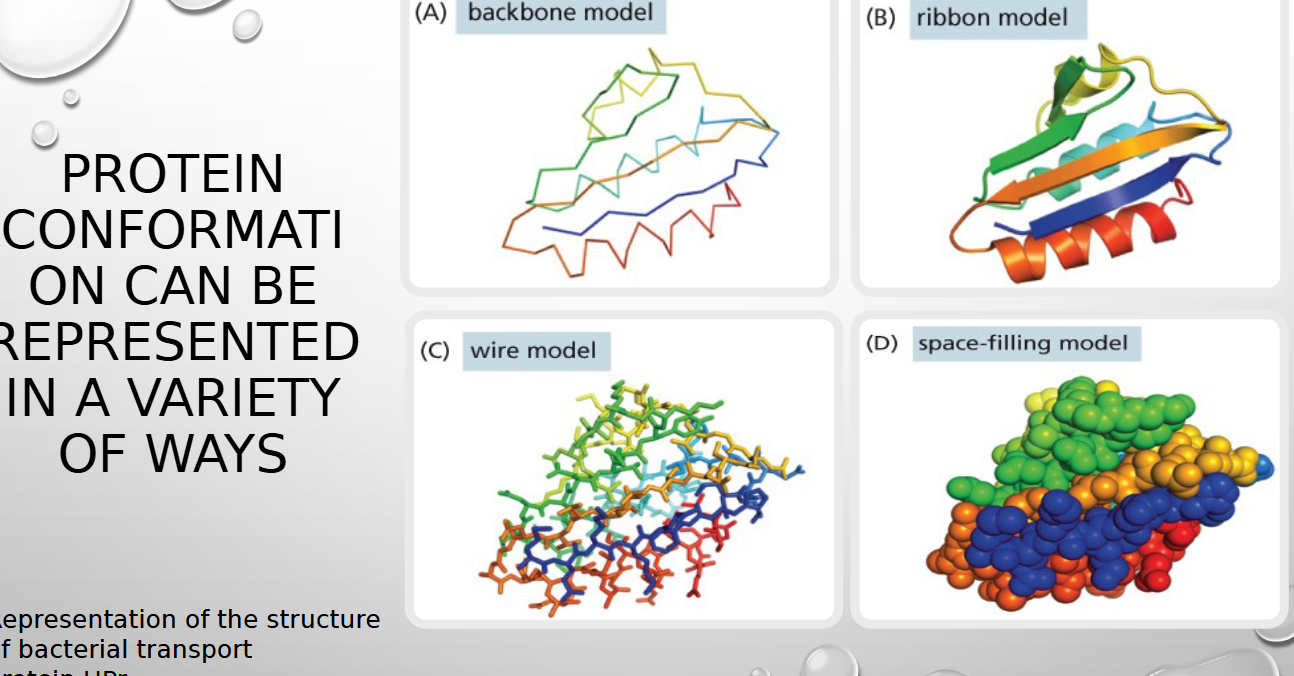
4 ways proteins are modeled (BWRS)
backbone, wire, ribbon, space filling model
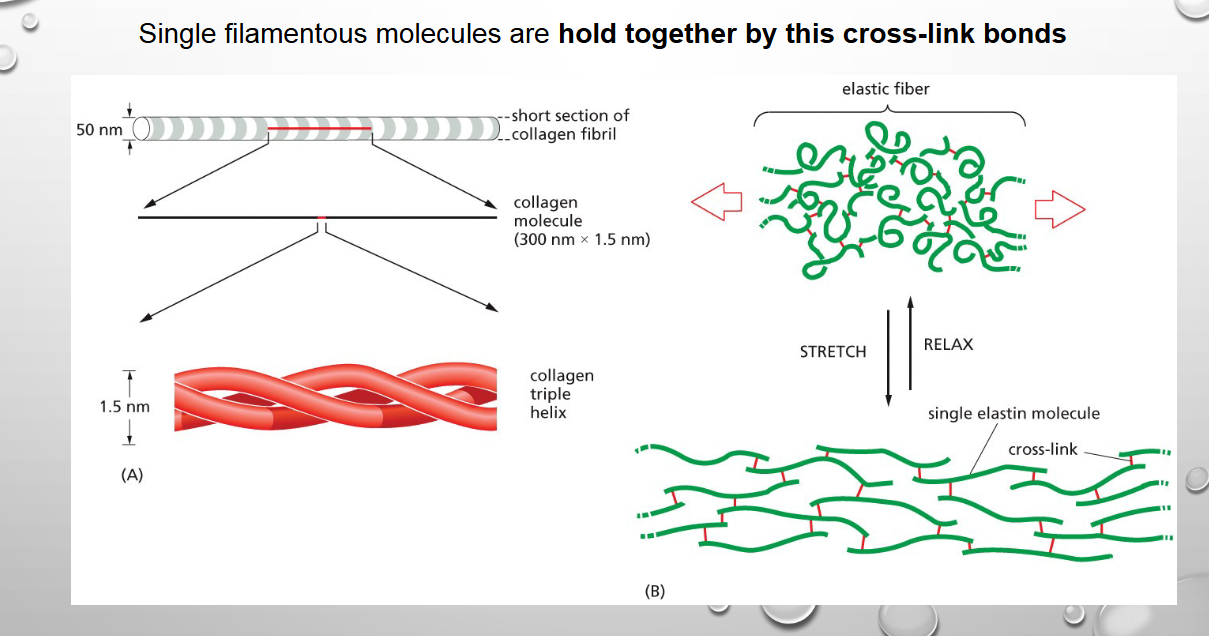
Single filamentous molecules are held together by
cross-link bonds
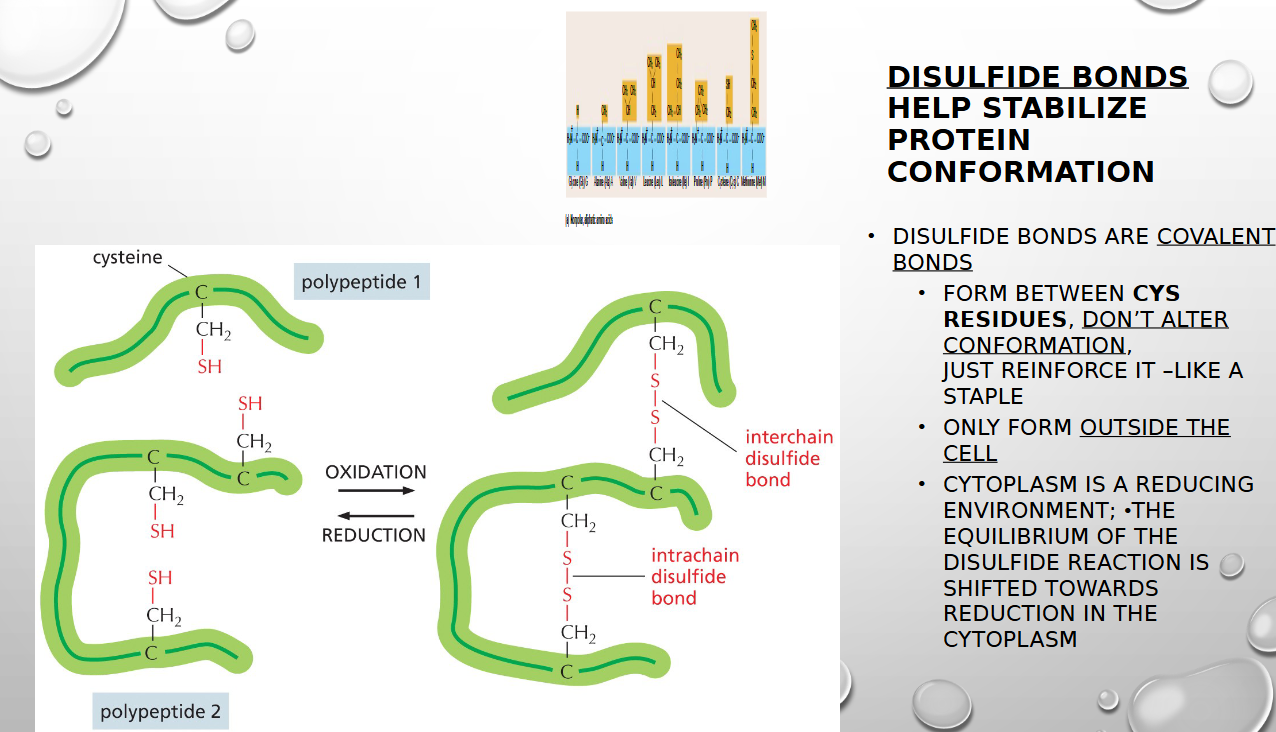
how do disulfide bonds stabilize protein conformation
by forming a strong, covalent cross-link between two cysteine amino acids, acting as a "staple" to hold different parts of a protein's structure together (only form OUTSIDE the cell)
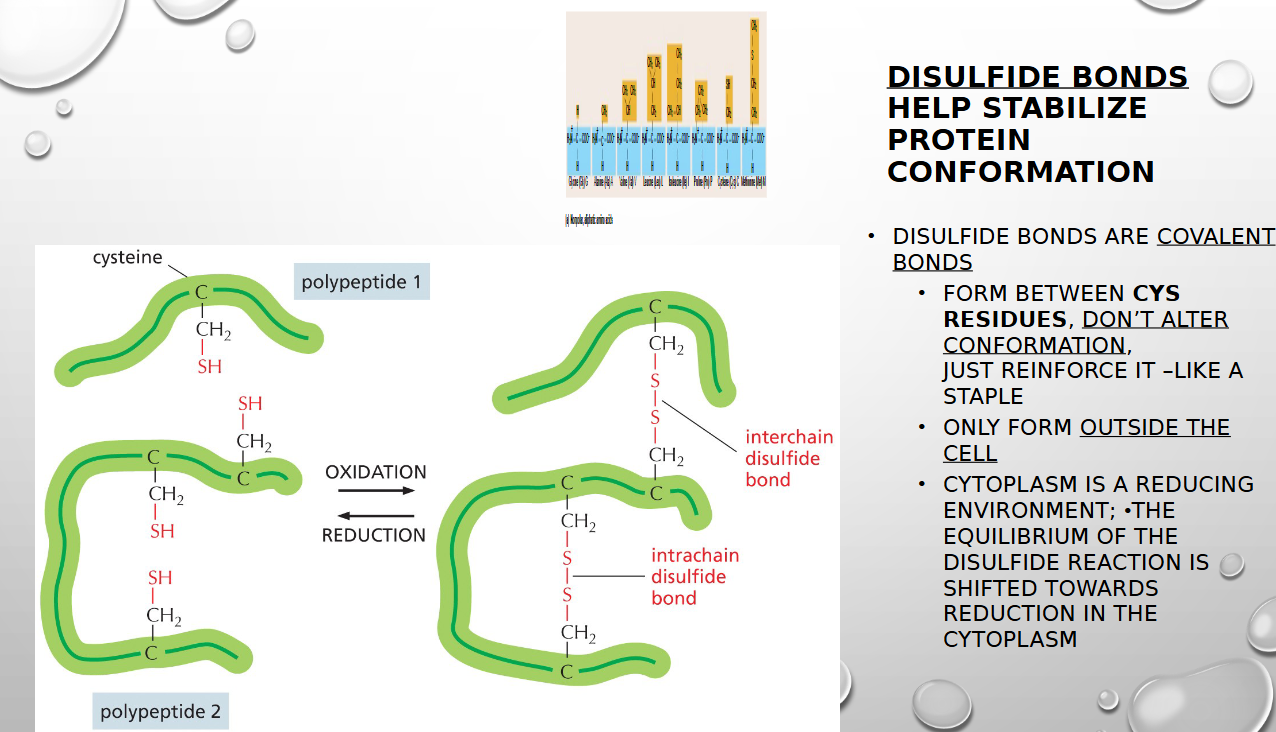
why do disulfide bonds stabilize protein conformation outside the cell only
the cellular environment inside the cytoplasm is reducing, which prevents protein formation. Outside, the environment is oxidizing so that favors the creation of the disulfide bonds
THE SHAPE OF A PROTEIN IS SPECIFIED BY ITS
AMINO ACID SEQUENCE
PROTEINS FOLD INTO A CONFORMATION OF LOWEST ENERGY THROUGH WHAT TYPE OF BONDS
NON-COVALENT BONDS (HYDROGEN BONDS, ELECTROSTATIC ATTRACTION, VAN DER WALLS FORCES, AND HYDROPHOBIC FORCES
protein four levels of organization
primary, secondary, tertiary, quaternary
THE Α HELIX AND THE Β SHEET ARE COMMON FOLDING PATTERNS BECAUSE
BECAUSE THEY INVOLVE STABLE BACKBONE ATOM BONDING
EXTRACELLULAR PROTEINS ARE OFTEN STABILIZED BY COVALENT CROSS-LINKAGES. ONE IMPORTANT CROSS-LINK IS THE
DISULFIDE BOND (S-S) THAT REINFORCE A PROTEIN (MOLECULAR STAPLER)
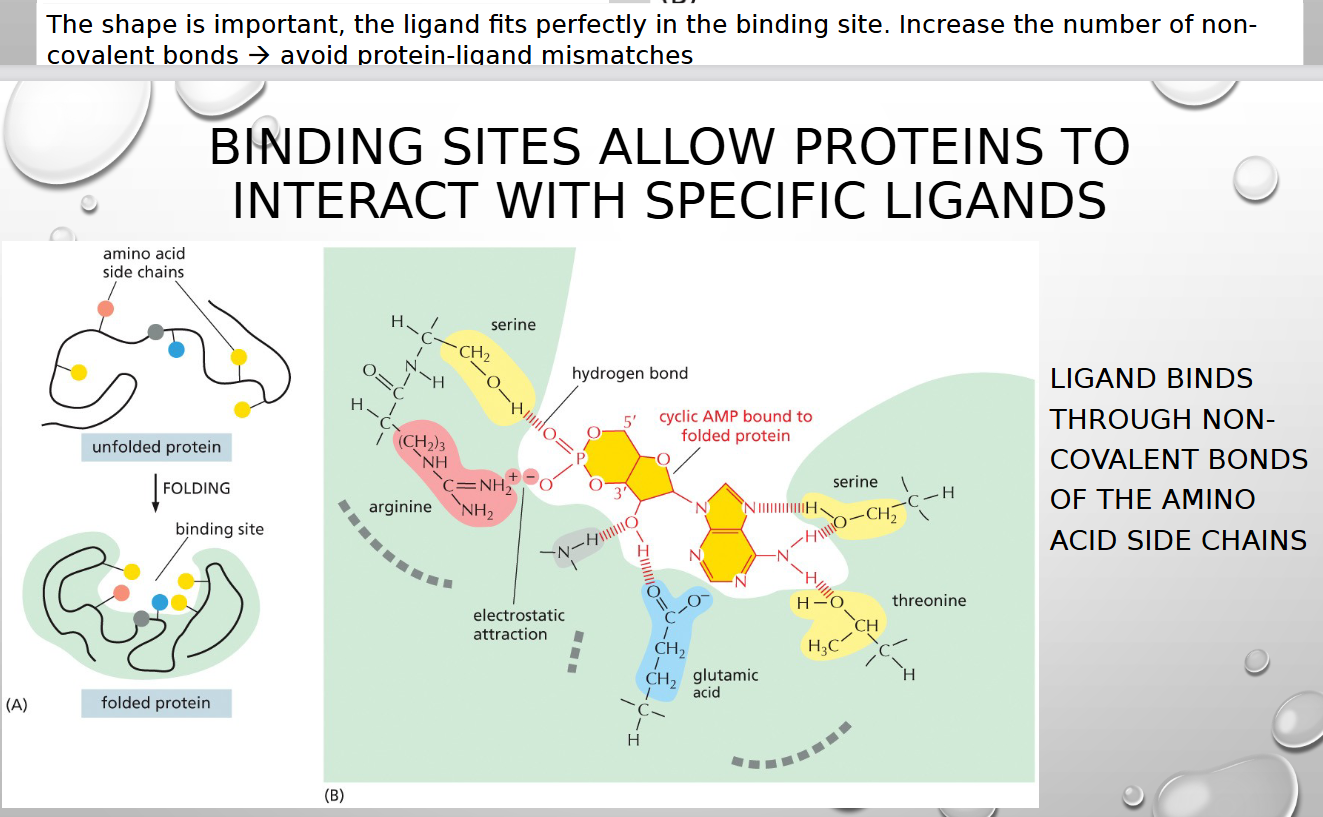
HOW DO PROTEINS WORK IN GENERAL
BY BINDING TO ANOTHER MOLECULE
ligand
substance, such as an ion or molecule, that binds to a larger target molecule
how do ligands bind to larger molecule (type of bond)
through non covalent bonds of amino acid side chain
ENZYMES ARE GROUPED INTO FUNCTIONAL CLASSES BASED ON
THE CHEMICAL REACTIONS THEY CATALYZE AND EACH ARE HIGHLY SPECIFIC TO A CERTAIN SUBSTRATE
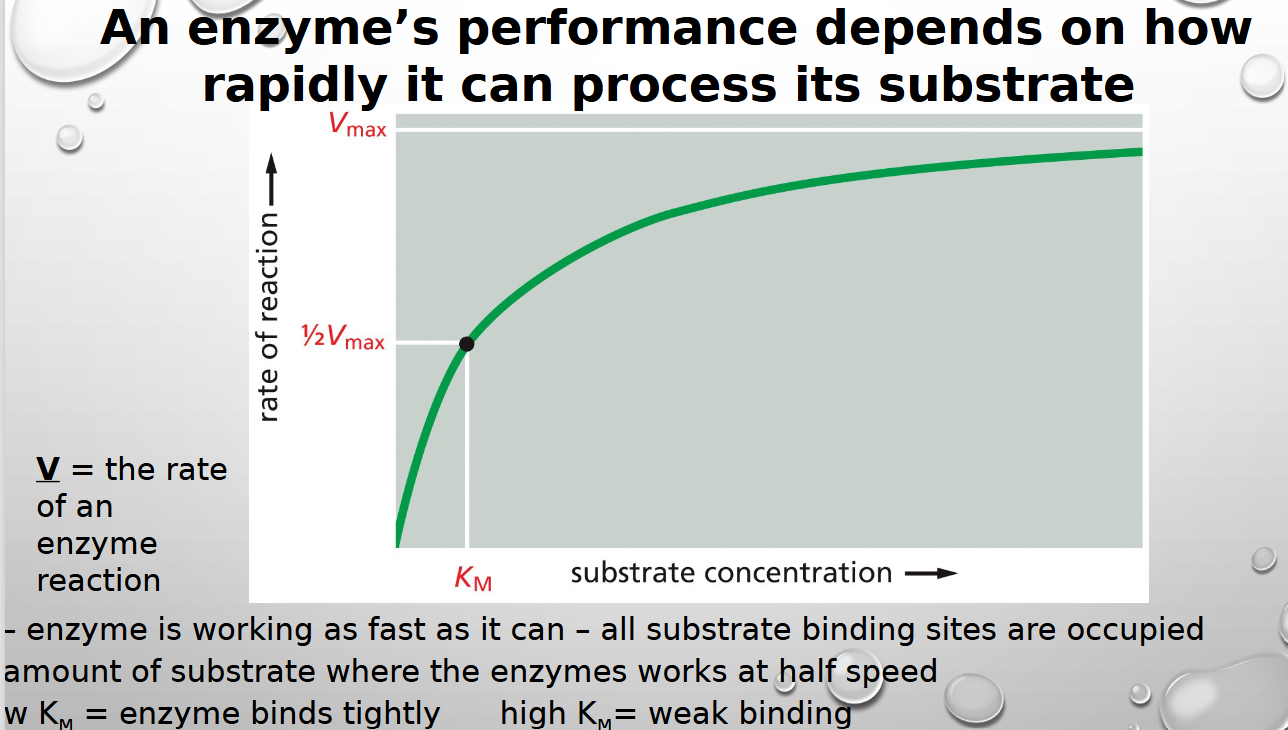
An enzyme’s performance depends on how rapidly it can process its
substrate
what is v and km in relation to enzymes
v is rate of enzyme reaction and km is how loosley it binds (HIGH KM IS WEAK)
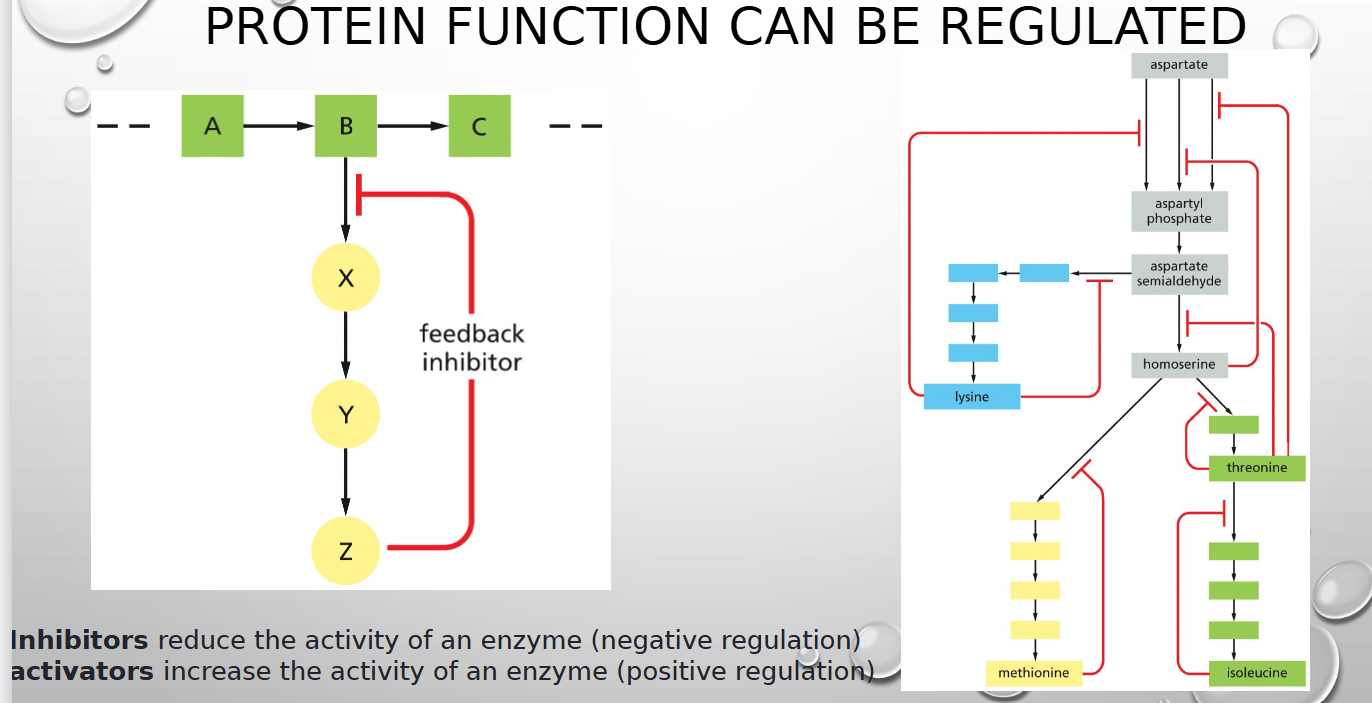
how are proteins controlled
feedback inhibition and positive regulation
feedback inhibition
a form of enzyme regulation whereby products prevent product formation
positive regulation
a mechanism, often an activator protein, that increases the frequency, rate, or extent of a biological process, such as gene expression
inhibitor vs activator
reduce vs increase activity of enzyme (neg vs pos regulation)
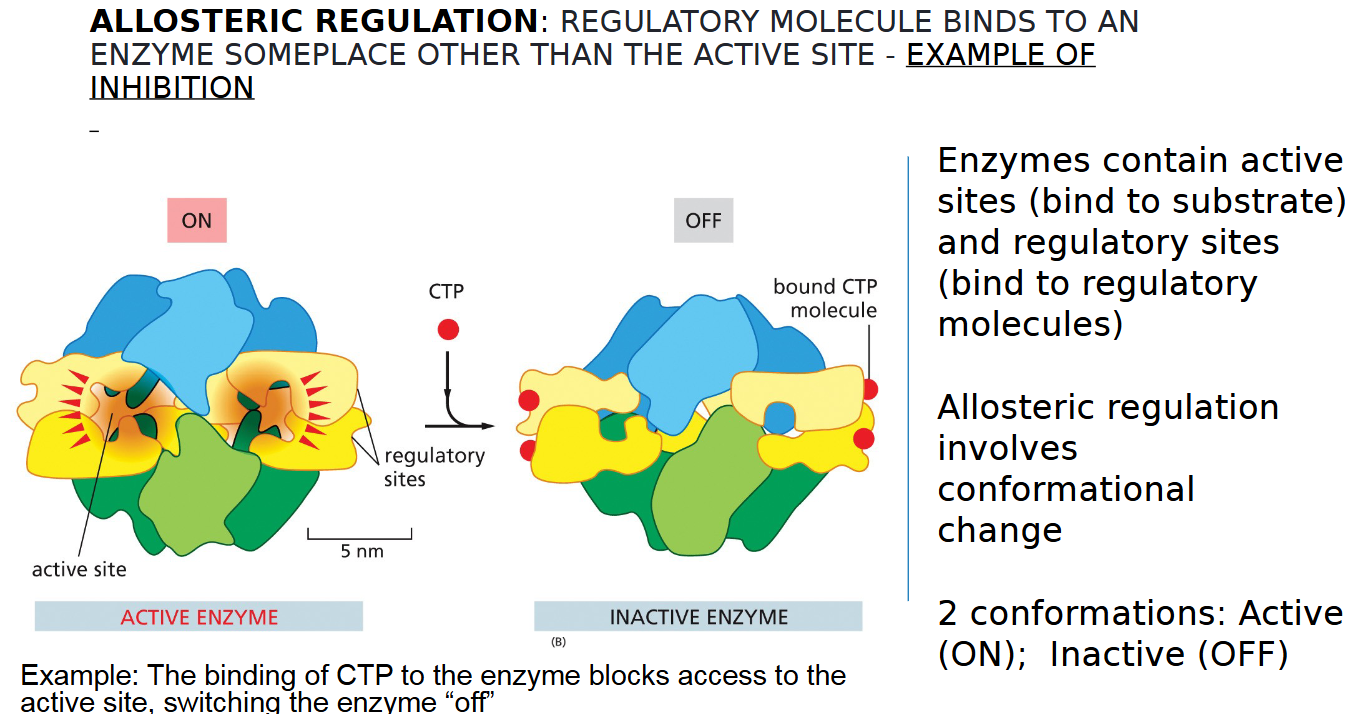
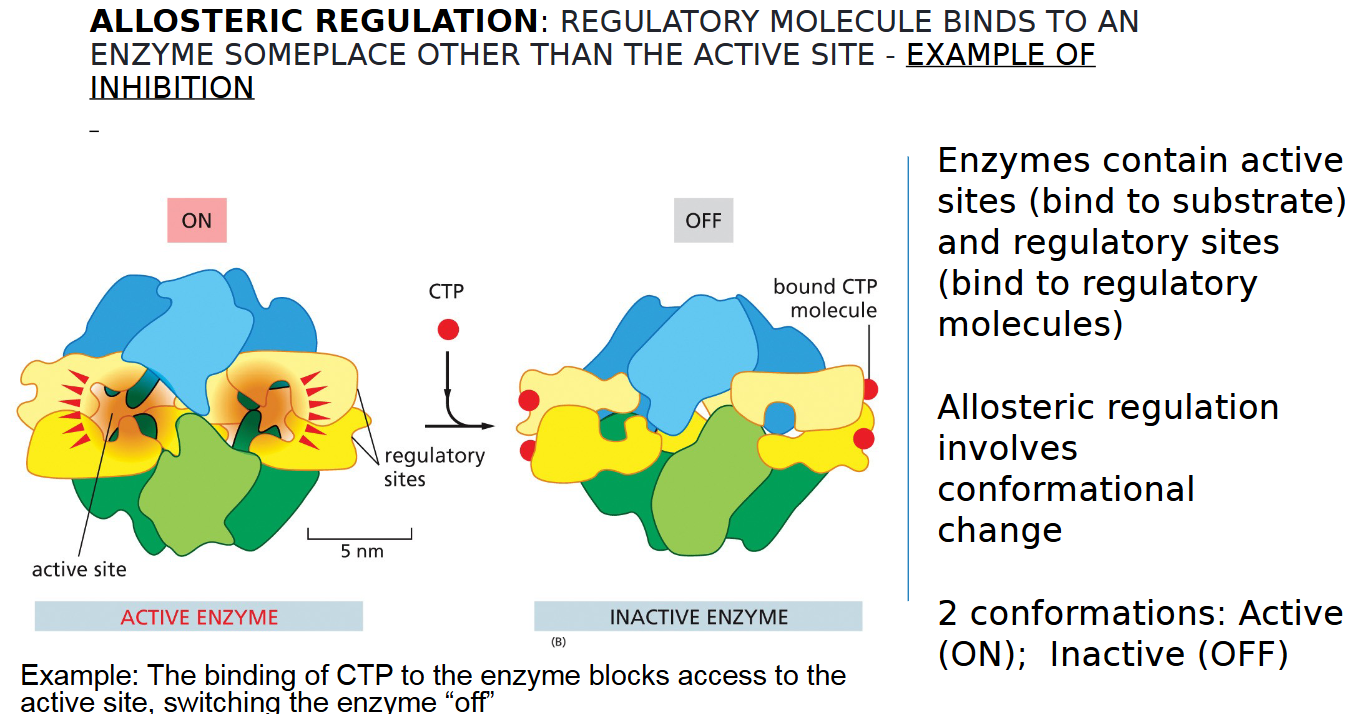
allosteric regulation inhibition example
REGULATORY MOLECULE BINDS TO AN
ENZYME SOMEPLACE OTHER THAN THE ACTIVE SITE TO TURN IT OFF
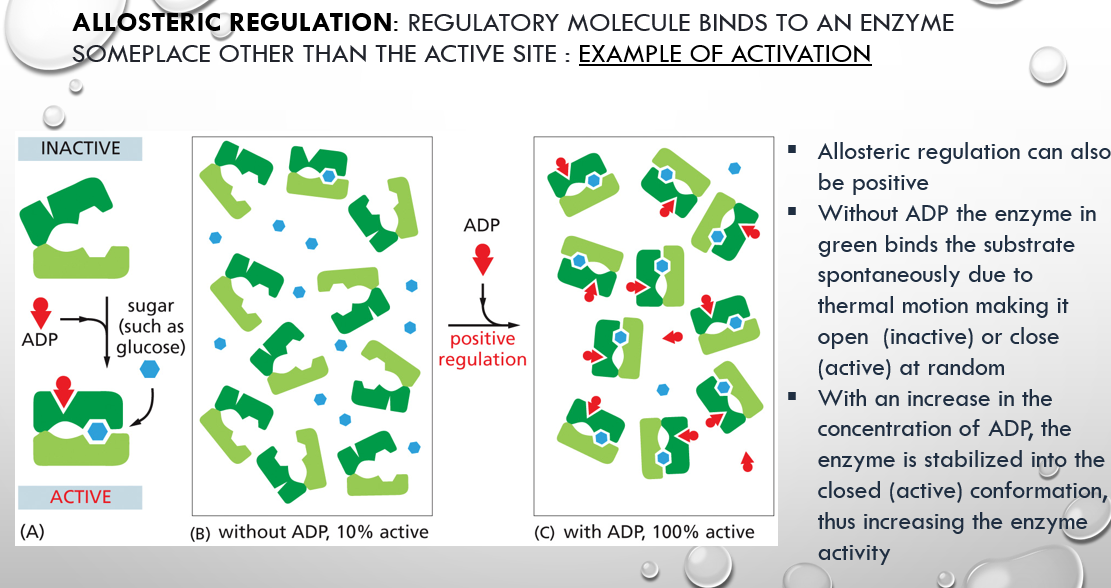
allosteric regulation activation example
more adp makes the enzyme in green more stabilized by keeping it closed on the substrate
two types of sites on an enzyme
inactive and active
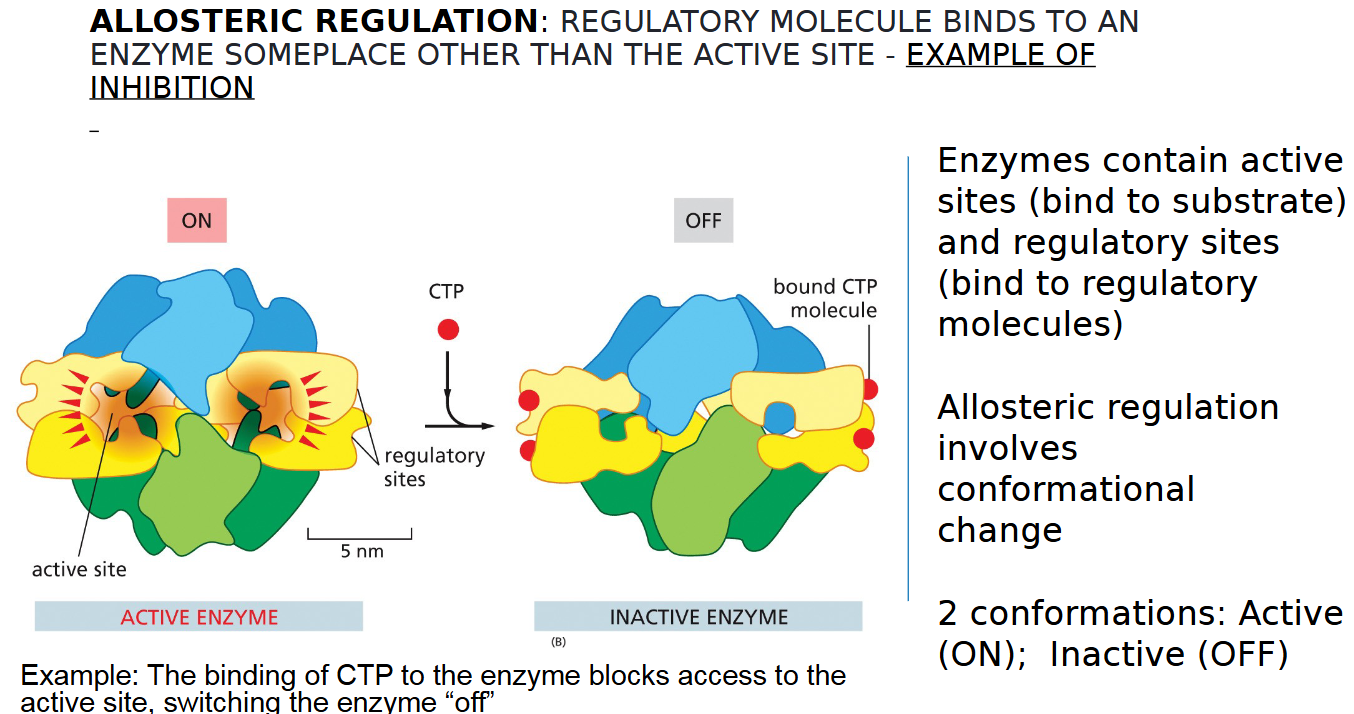
2 conformations of allosteric regulation
active vs inactive
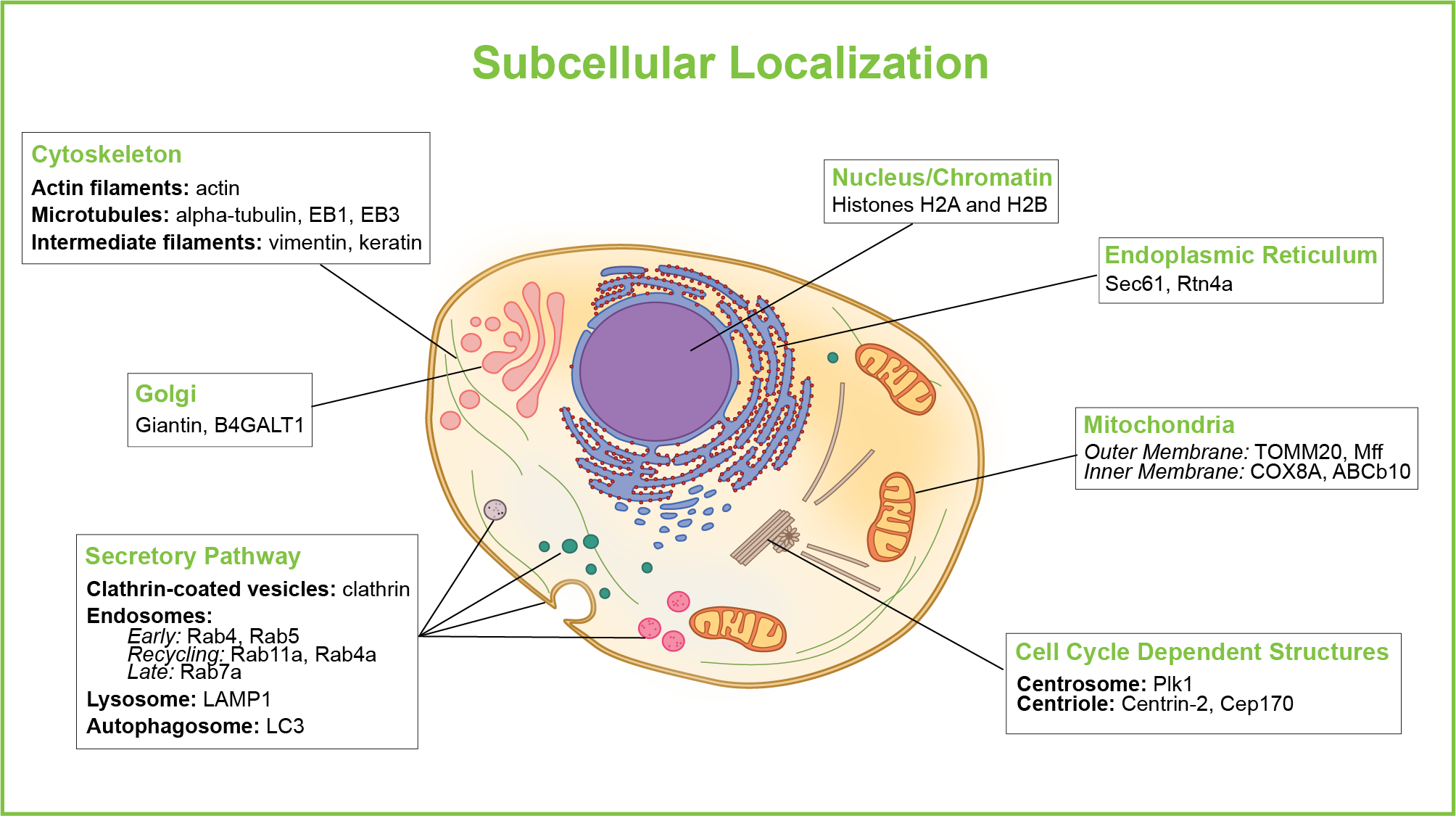
protein localization
process by which proteins are delivered to and maintained in specific locations within a cell or organism
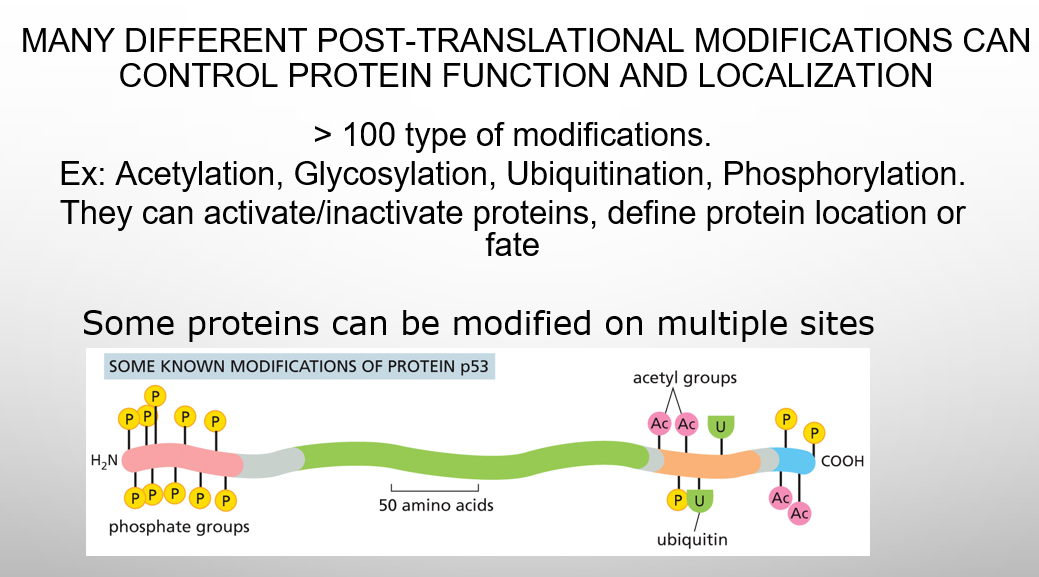
what can control protein function and localization
different post-translational modifications
Phosphorylation can control protein activity by causing
conformational change
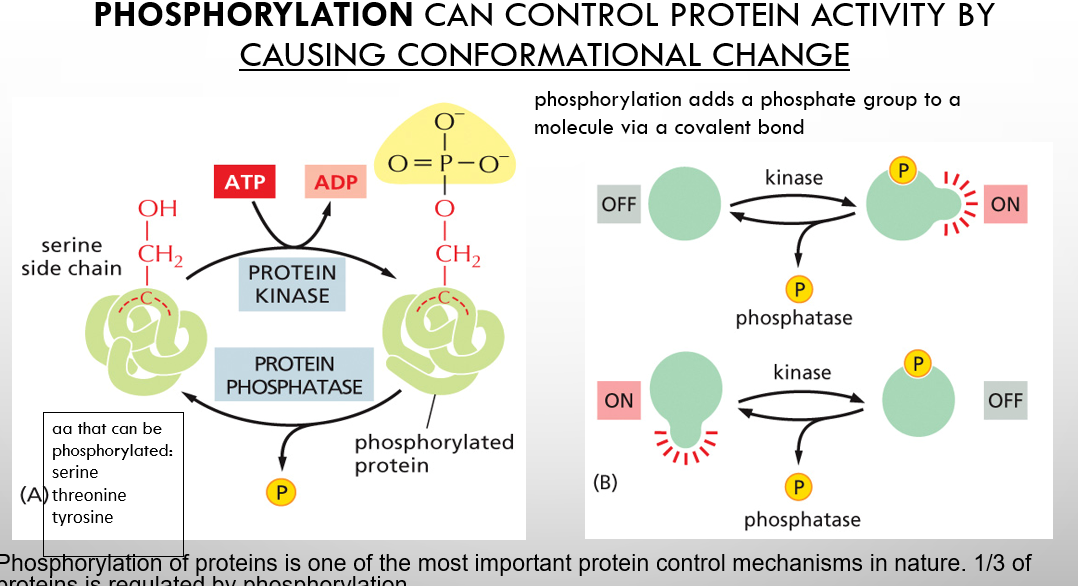
what chemical group does phosphorylation add to a molecule and how
phosphorylation adds a phosphate group to a molecule via a covalent bond
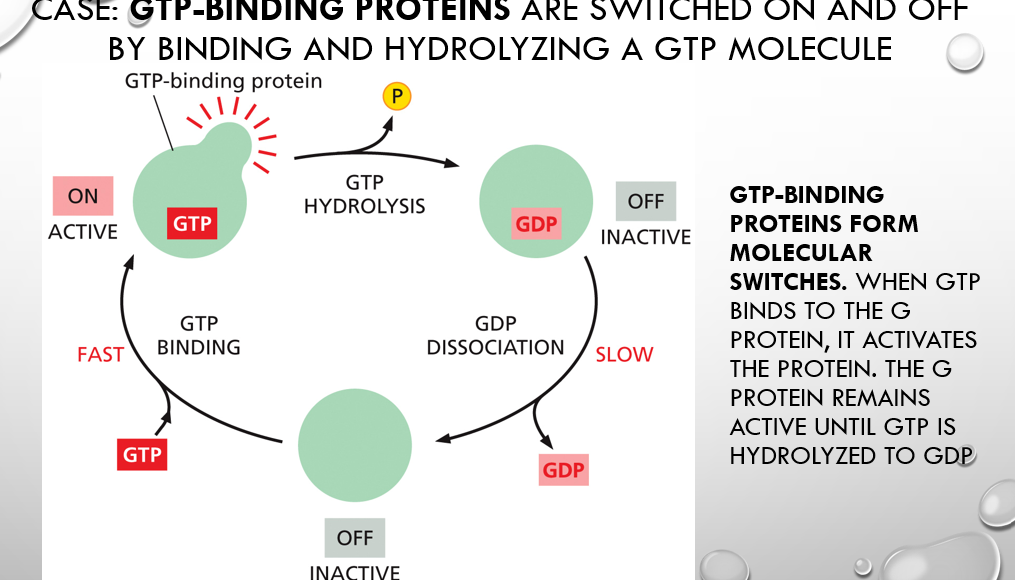
how do gtp binding proteins form molecular switches
When GTP binds to the G protein, it activates the protein. The G protein remains active until GTP is hydrolyzed to GDP
atp hydrolysis allows for what
Atp hydrolysis allows motor proteins to produce directed movement in cells
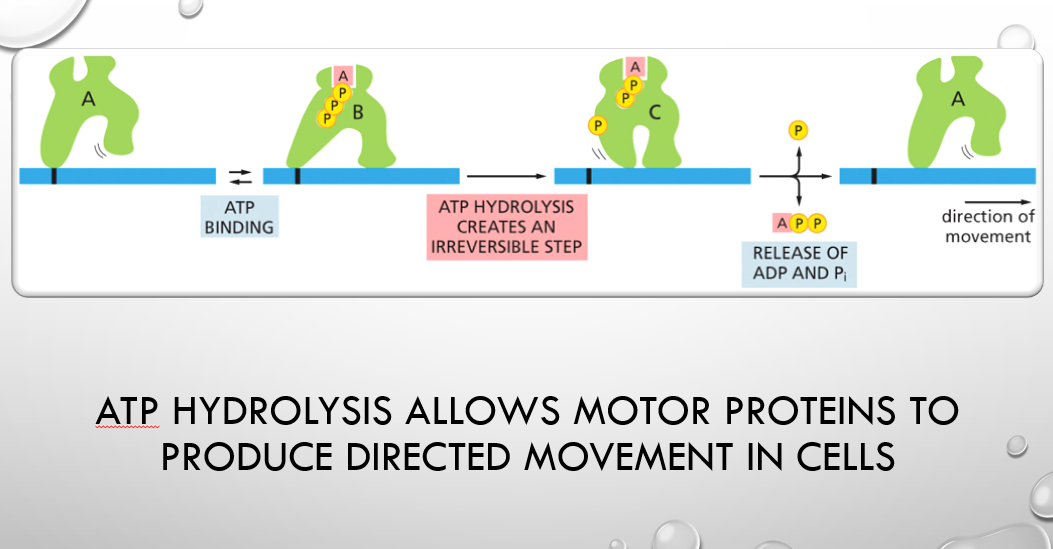
The Catalytic Activities of Enzymes Are Often Regulated by Other Molecules called
inhibitors or activators
four types of enzyme regulation (AEPG)
allosteric, enzyme modification, phosphorylation, gtp binding
Allosteric regulation involves the binding of what to what
regulatory molecules (inhibitors or activators) to the enzyme. Involves enzyme conformational change
Enzyme modification has around how many types
> 100 type of modifications. Ex: Acetylation, Glycosylation, Ubiquitination. Most used: Phosphorylation, which Can Control Protein Activity by Causing a Conformational Change
Phosphorylation involves the addition of what to what
phosphate group (PO₃²⁻) to a Protein. This process is a reversible post-translational modification that changes the protein's shape, activity, and ability to bind other molecules, acting as a molecular switch.
GTP binding proteins, also known as G proteins, act as "molecular switches" because
When GTP binds to the G protein, it activates the protein. The G protein remains active until GTP is hydrolyzed to GDP
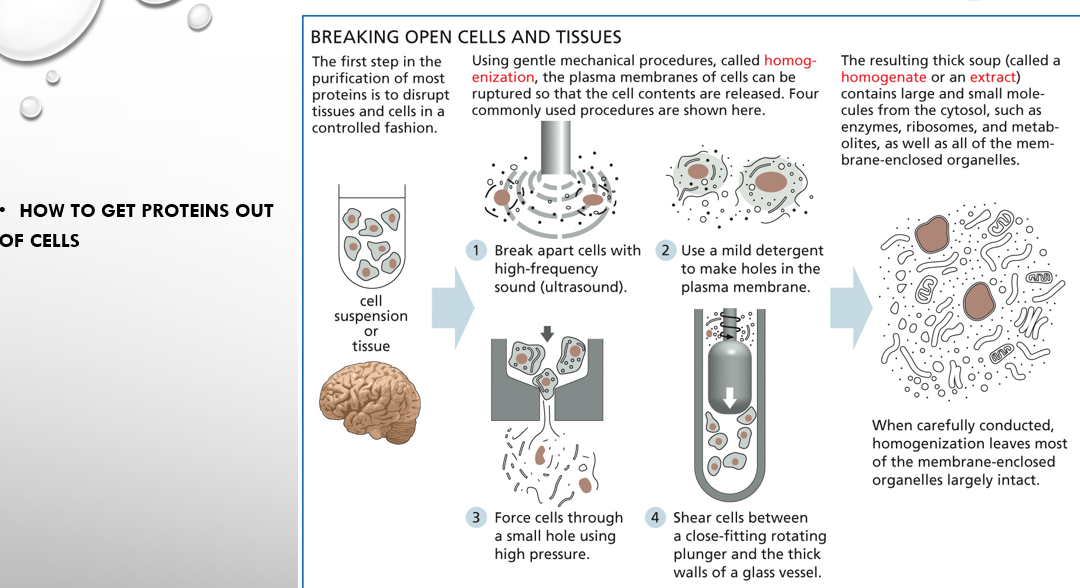
how to get proteins out of cells (four steps)
break them apart with high ultrasound frequency
use detergent to make holes in them
force cells through small holes
use plunger and tube to smash them up
homogenate vs supernatant vs pellet of centrifugation
homogenate is the mixture before centrifugation while supernatant is the less dense part of the mix and pellet is the more dense part
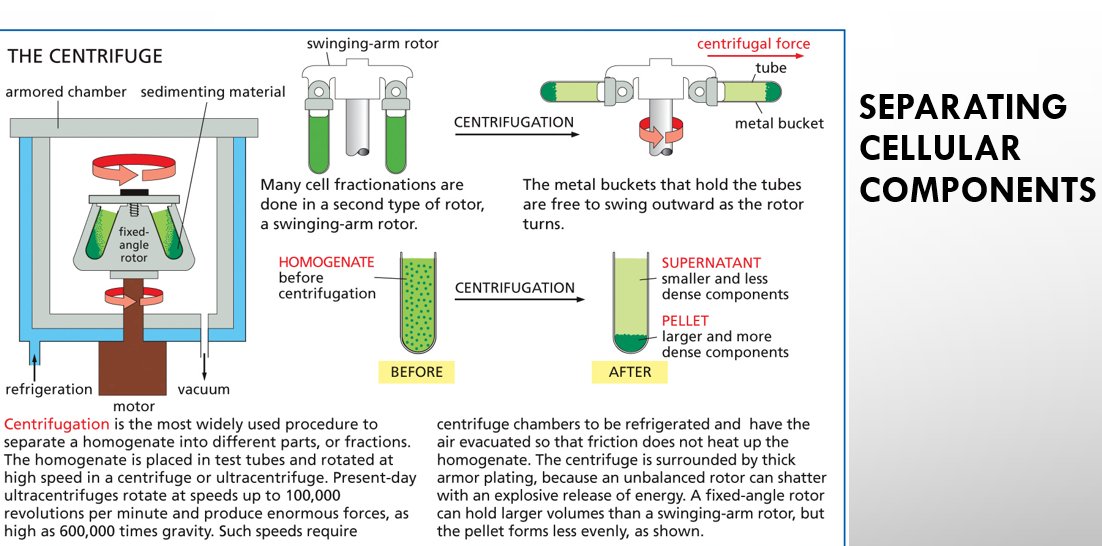
centrifugation vs ultracentrifugation
Centrifugation separates components of a mixture by spinning, while ultracentrifugation is a specialized form of centrifugation using much higher speeds to separate smaller, more delicate particles
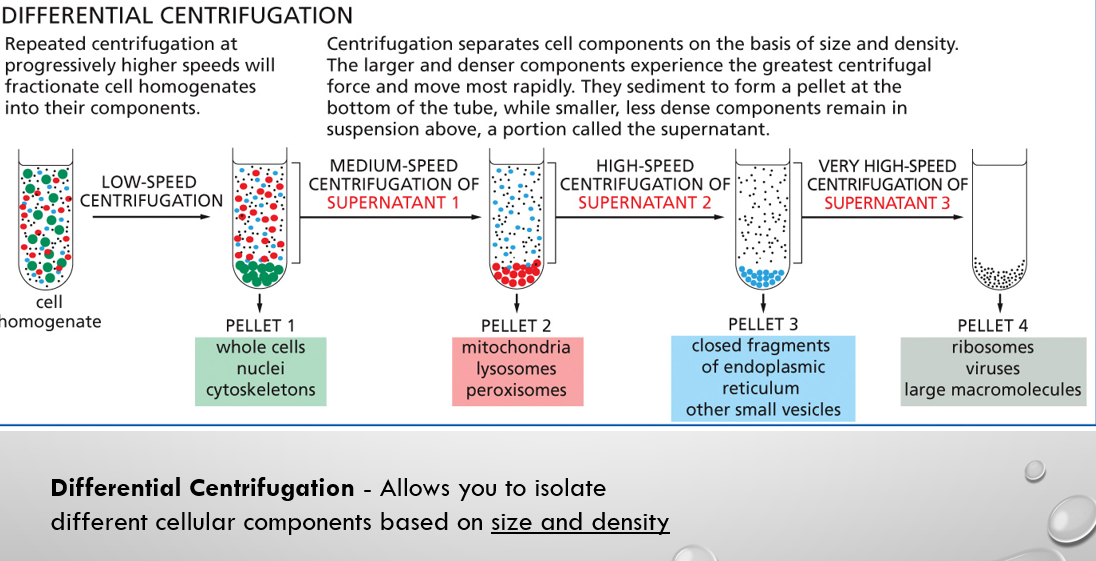
differential centrifugation
a specific type of centrifugation that uses successive increases in centrifugal force to separate particles based primarily on their SIZE AND DENSITY
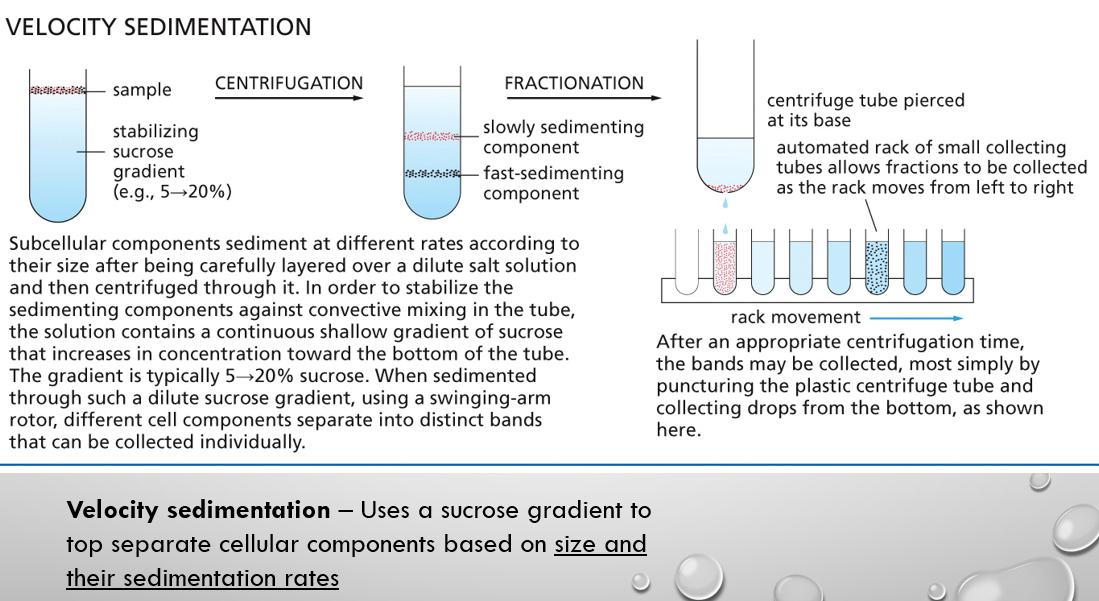
velocity sedimentation
centrifugation with a sucrose gradient to top separate cellular components based on size and their sedimentation rates
equilibrium sedimentation/density gradient centrifugation
Allows you to isolate different cellular components based on their buoyant density
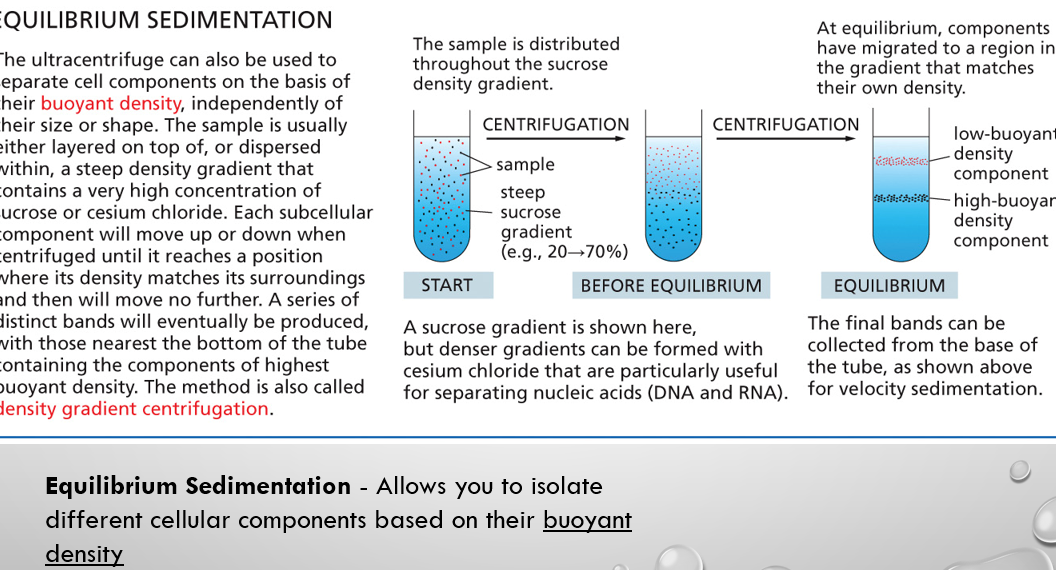
velocity sedimentation vs equilibrium sedimentation biology
separates molecules based on how fast they move (their sedimentation rate) through a solution in a centrifugal field vs. separates molecules based on their buoyant density. Centrifugation continues until molecules stop moving and settle at a certain point where their density equals that of the surrounding gradient.
protein column chromatography
uses resin-filled columns to separate and purify proteins from complex mixtures based on their unique properties, such as size, charge, and specific binding affinities
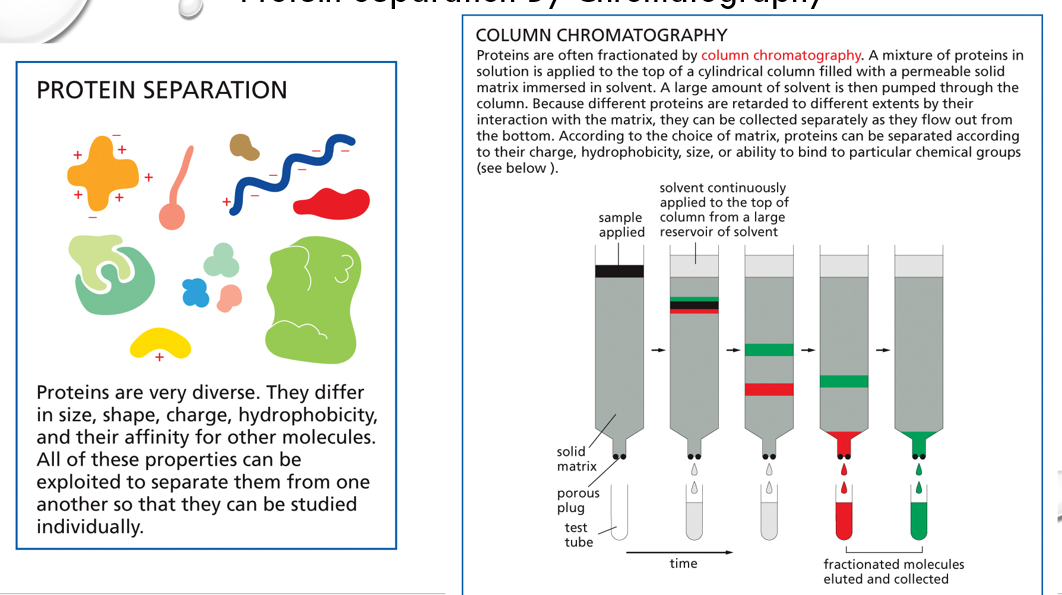
3 types of chromatography (GAI)
gel, affinity, ion
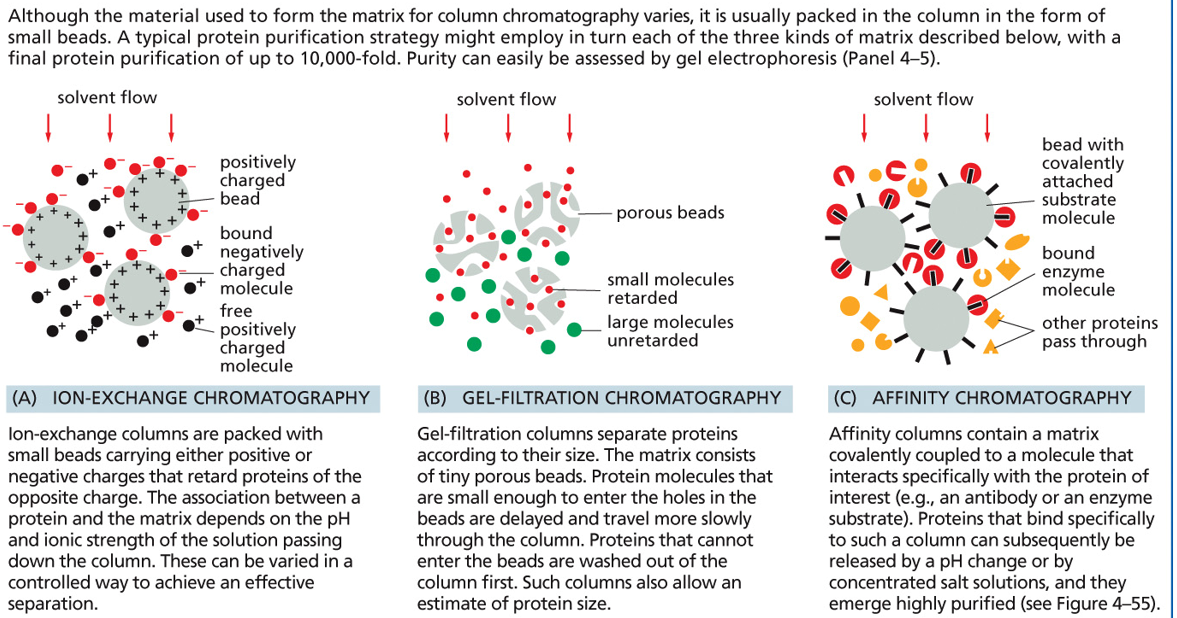
ion exchange chromatography
separates proteins by charge. beads have charges and oppositely charged proteins stick, while same-charged proteins flow through
gel chromatography
separates proteins by size. Beads have pores, and small molecules get trapped in pores (move slower). Large molecules can’t enter pores (move faster)
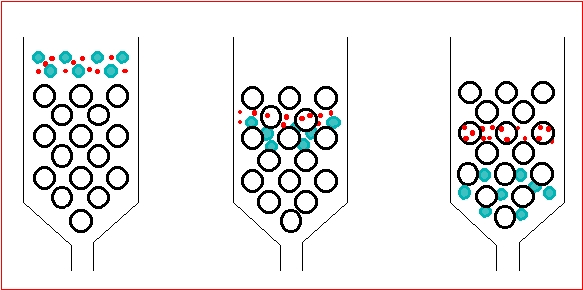
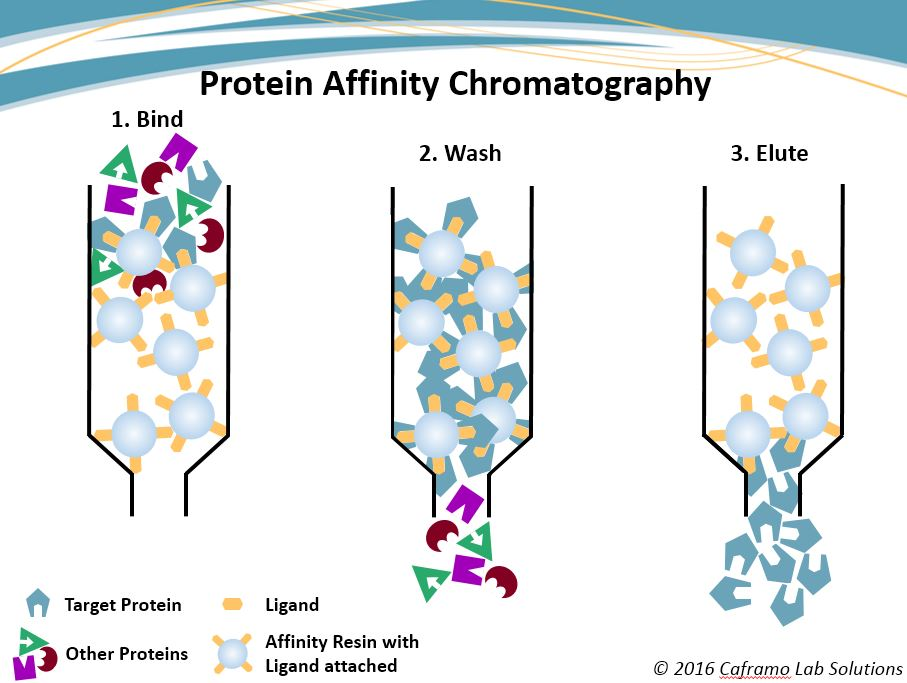
affinity chromatography
separates proteins by specific binding. Beads have a molecule (e.g., antibody or substrate) that binds only the target protein. Other proteins wash away. Bound protein is then released by pH change or salt
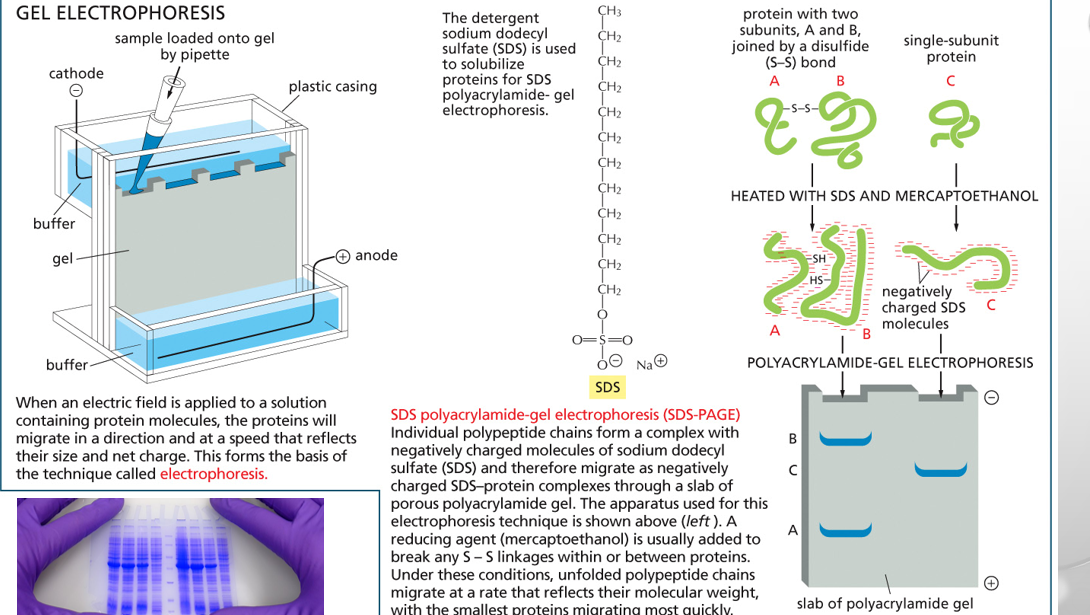
gel electrophoresis
uses an electric current to separate macromolecules like DNA, RNA, or proteins based on their size and electrical charge
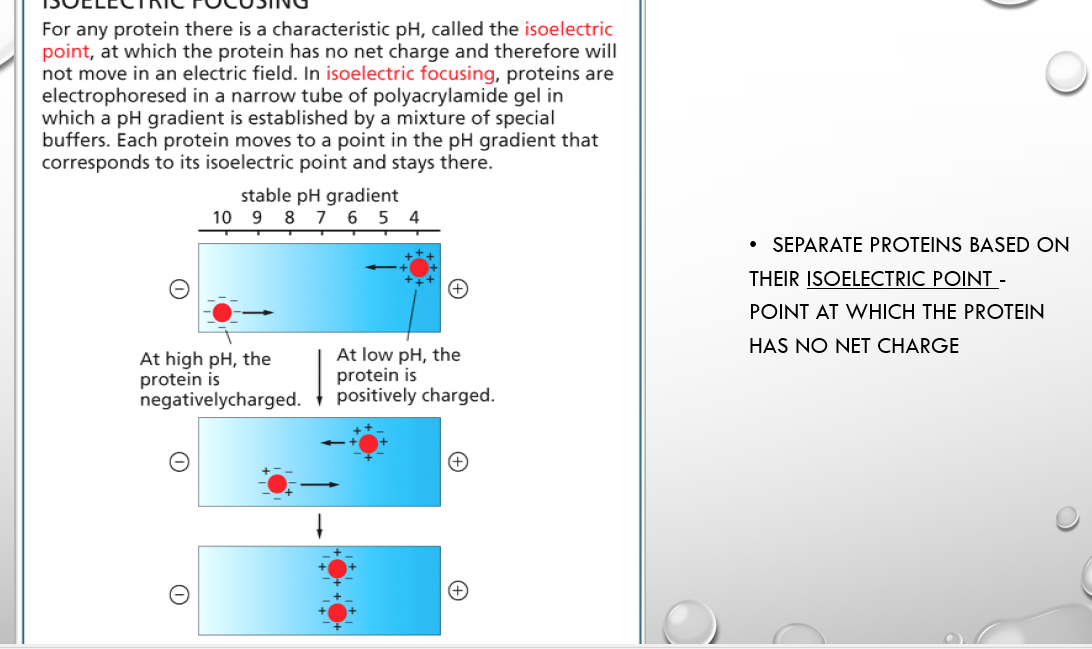
isoelectric focusing
separates proteins based on isoelectric point where they are at a pH with no net charge and will thus not move in an electric field
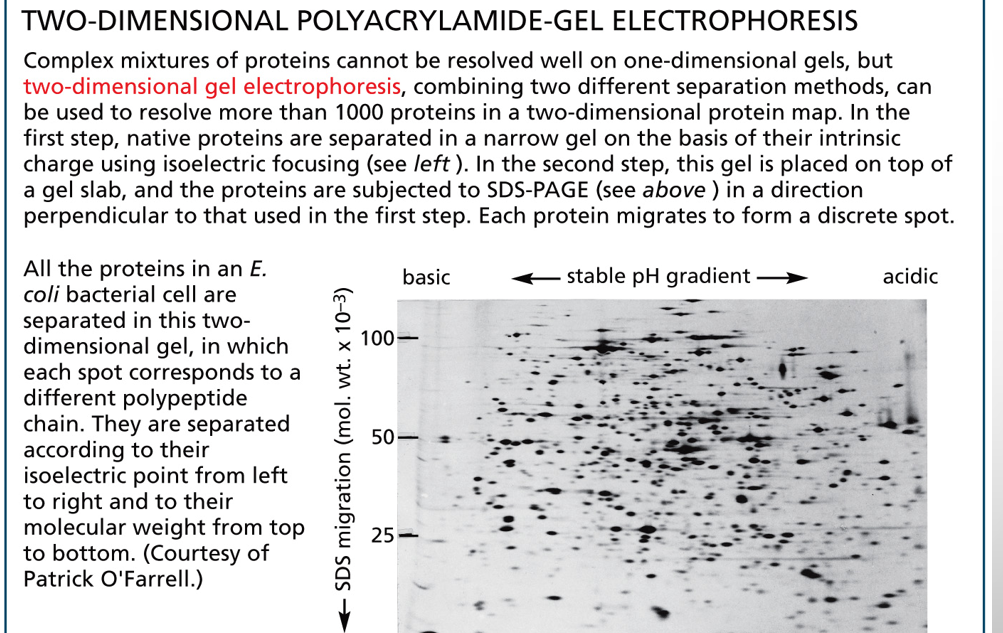
two dimensional gel electrophoresis
separates complex protein mixtures into thousands of distinct "spots" on a single gel by first separating proteins by their isoelectric point (charge) and then by their molecular weight (size)
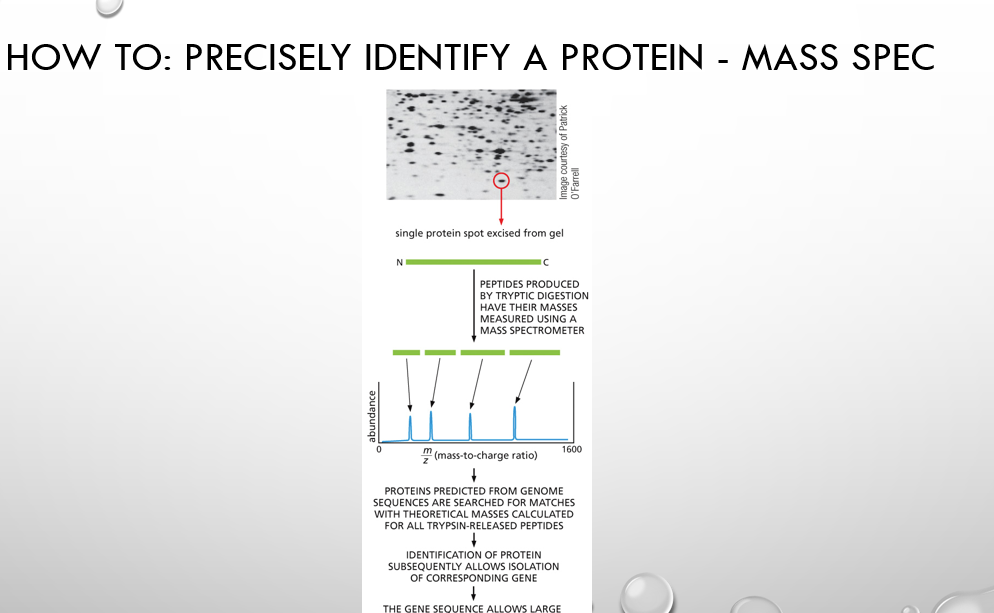
how to identify a protein with mass spec steps (CCPC)
Cut the protein from a gel.
Chop it into small pieces (peptides) with an enzyme.
Put peptides into a mass spectrometer → it measures their sizes (mass/charge).
Compare sizes with a database.
Match found = protein identified.
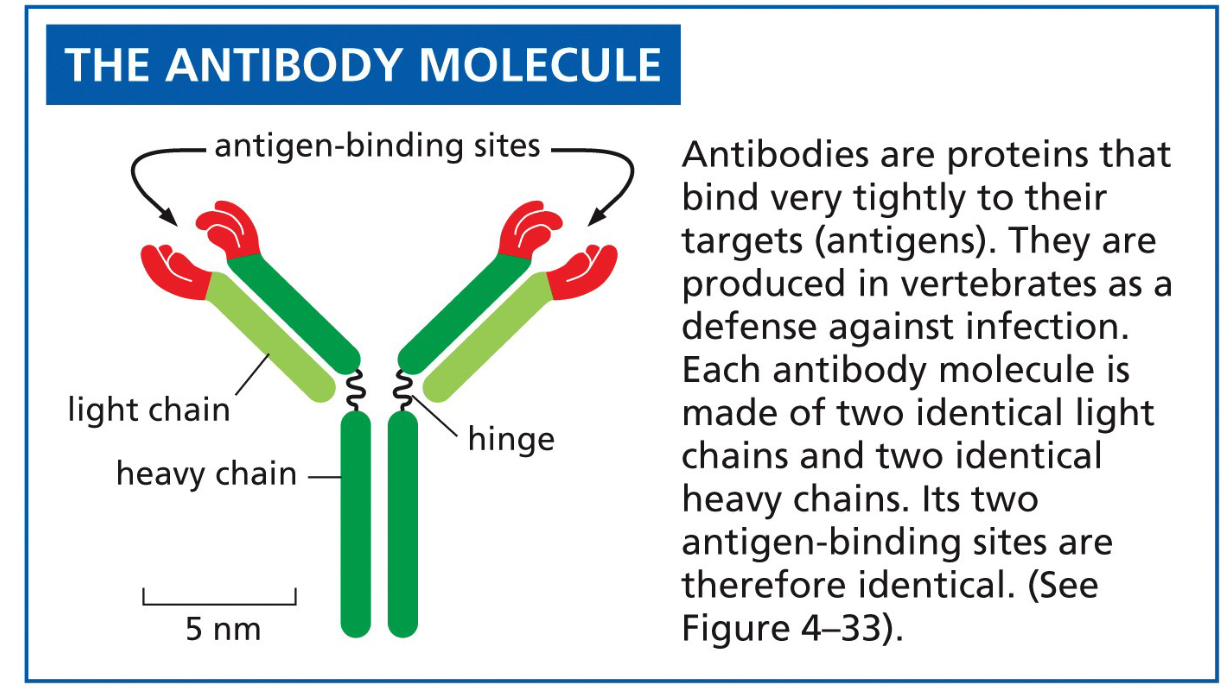
what are antibodies
proteins produced by immune system to bind very tightly to their targets (antigens)