2020 L8 Protein Degradation
1/23
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
24 Terms
Degradation Pathways
Ubiquitin Proteasome System (UPS)
Autophagosomal-Lysosomal Pathway (ALP)
Both are very enzymatically selective
Pathway Determination
Critical determinant = substrate size
UPS degrades single polypeptides, can fit into narrow channel of the proteasome
APL degrades larger structures e.g. protein aggregates, organelles or pathogens
UPS
Major pathway for protein degradation, 80% of protein turnover
Driven by ubiquitin as a degradation marker - ubiquitin is conjugated to a protein, signal that protein needs to be degraded.
Posttranslational modification - protein’s function altered after being synthesised
Proteins tagged with ubiquitin have short half-lives (minutes to hours)
Central to the UPS is the cytosolic 26s proteasome
26S Proteasome
Large multi-subunit protease complex
Composed of two subunits
20S core protease houses peptidase activity
19S regulatory particle - binds to core protease in ATP presence, contains ATPase domains
Ubiquitin-meidated degradation
Ubiquitin-tagged substrates bind to regulatory particles, activating ATPase domains
Conformational change following ATP hydrolysis allows access to the core protease
Ubiquitin residues are removed and recycled, protease cleaves protein e.g. trypsin-like, chymotrypsin-like and caspase-like
Ubiquitin
Small protein, contains C-terminal glycine - the site of ubiquitin attachment
Conjugated to lysine residues in proteins e.g. Lysine 6, 11, 27 etc.
Ubiquitin tags are diverse and dictate the outcome
Polyubiquitination (particularly of lysine 11 and 48) is most potent signal for degradation because it recruits shuttle factors
Monoubiqutination alters protein localisation and conformation
Ubiquitin cascade
Conjugation occurs via enzyme cascade
Ubiquitin activated by ATP, becomes bound to E1
Ubiquitin transferred to E2 and finally to E3
E3 enzymes are ubiquitin ligases, conjugate ubiquitin to a target protein
Additional Roles of UPS
Small fraction of proteins are immediately targeted for degradation during translation
DRIP (defective ribosomal product hypothesis) -hypothesised to provide source of proteins for MHC-I presented antigenic peptides
Ubiquitination occurs at stalled ribosomes, degrading nonsense mRNA and no-go decay proteins
UPS Molecular Chaperones
Molecular chaperones: stabilise misfolded proteins in a non-aggregated state - proteasome cannot degrade large proteins and aggregates
E3 ligase CHIP interacts directly with Hsp70 and 90 via its tetratricopeptide domain
Hsf1 upregulates several E3 ligases
Hsf1 is ubiquitinated following proteotoxic stress
Endoplasmic Reticulum Associated Degradation (ERAD)
Transports misfolded proteins from ER to cytosol
E3 ligase Hrd1 ubiquitinates ER proteins, has a transcolon function
Valosin (ubiquitin binding factor) contains protein VCP/p97 - transports proteins from ER to cytosol to be degraded
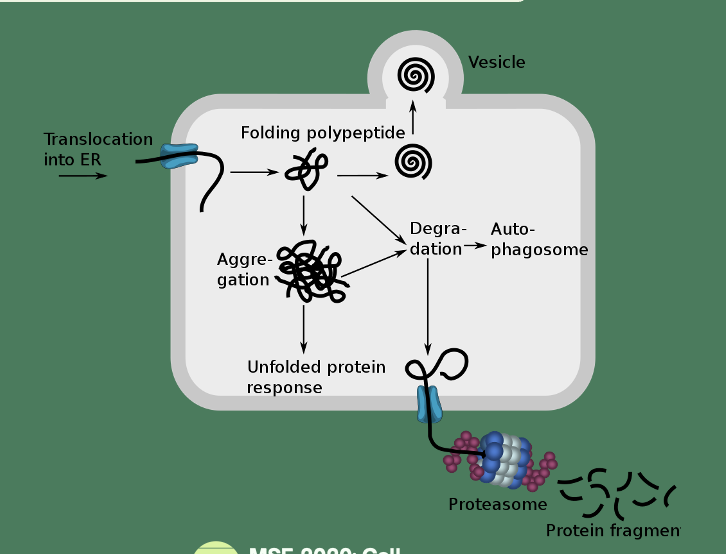
Cystic Fibrosis (CF)
Lethal autosomal recessive disease caused by mutations in Cystic Fibrosis Transmembrane Conductance Regulator (CTFR) gene
Common in Europeans, 1 in 25 are carriers
Associated w respiratory infection due to thickened mucus - CTFR transports chloride ions
CF Mutations
Characterised by allelic heterogeneity - different mutations at the same gene locus can cause the same disease or phenotype. In cystic fibrosis, multiple mutations in the CFTR gene can lead to similar clinical symptoms.
Compound Heterozygosity - genetic condition in which an individual inherits two different mutations in the same gene, one from each parent. In cystic fibrosis, this means a patient may have two different mutations in the CFTR gene that both contribute to the disease.
CTFR undergoes co- and post-translational folding and core glycosylation in ER
Fully glycosylated in the Golgi and inserts in the apical membrane
F508Δ
Deletion of phenylalanine at position 508 - common mutation of CTFR
Nascent peptides misfolds and is immediately targeted to the proteasome for degradation
No functional CTFR is localised to the membrane
Chloride transport is inhibited + Na transport into the cell through ENaC (epithelial Na channels)
Juxtanuclear Quality Control Compartment (JUNQ)
Centre of ubiquitination and degradation of UPS in response to cell - only one per cell
Ubiquitinated proteins are sequestered into the JUNQ
Protein used to be thought as a random process, but recent studies w fluorescent microscopy shows protein aggregation is tightly regulated - sequestered into inclusion bodies
Many chaperone and proteasome complexes in JUNQ, concentrates proteins for degradation
@ mitosis mother cell retains JUNQ through asymmetrical inheritance - therapies targeting this mechanism could mitigate neurodegenerative diseases by preventing the buildup of toxic aggregates that contribute to neuronal death.
ALS
Eliminates large protein aggregates, protein complexes, organelles, pathogens
Best characterised form: macroautophagy, where double membrane structure autophagosome, engulfs cell material
Autophagy Activation
Macroautophagy initiated by ULK1 kinase, triggers a kinase cascade
ATG8 protein complexes (LC3 and GABARAP in higher eukaryotes) begin to assemble
ATG8 pcs bind to phophatidylethanolamine (phospholipid) on the phagophore
Phagophore matures into autophagosome, fuses with a lysosome
Ubiquitin, UPS and ALP
Many autophagy receptors contain a ubiquitin-binding domain, allowing ubiquitinates proteins/organellles to be processed by autophagy in addition to UPS
Significant crosstalk between UPS and ALP
Autophagy receptors e.g. p62 contain ubiquitin-binding domains, proteasome binding domains and LIR domains (phagophore)
ALP can compensate if UPS is overwhelmed - decision of which to use governed by avidity of protein complex
Heat shock can trigger high ubiquitin levels as UPS proceeds, which promotes autophagy
Lysosome
Organelles composed of acidic lumen and lysosomal membrane
Lumen is host to hydrolytic enzymes e.g. nucleases, proteases, phosphatases, lipases, sulfatases
A vacuolar H+ ATPase transports H+ into the lumen
Associated membrane proteins include SNARE proteins, LAMP1 and LAMP2
Glycocalyx
Composed of glycoproteins and glycolipids, covalently attached to membrane of lysosome
Protects the lysosome from attack by lytic enzymes in cytoplasm
Lysosomes in autophagy
During macroautophagy autophagosomes fuse with lysosomes through LC3 proteins
Molecular chaperones can also promote autophagy through CMA (chaperone-mediated autophagy)
Proteins recognised through KFERQ motifs by certain heat shock proteins e.g. Hsc70
LAMP2 assists in translocation of Hsc70-bound proteins into lysosomal lumen
Lysosomes in nutrient sensing
Rag-GTPases and Ragulator localise mTORC1 to the lysosomal membrane
mTORC1 central to protein, lipid and nucleotide synthesis, and regulates autophagy
mTORC1 senses amino acid conc. through Sestrin 2 and Castor - bind leucine and arginine respectively
high amino acids keep mTORC1 active
Inactivation of mTORC1 through amino acid deprivation causes ULK1 to become active
Organelle degradation
Driven by selective autophagy to remove defective organelles
Mitophagy (autophagy turnover of mitochondria) can be ubiquitin-dependant or independant
Pink1/Parkin tag outer mitochondria proteins with ubiquitin
Mitophagy during erythrocyte maturation is ubiquitin independant
Ribophagy is ubiquitin-dependant
ER autophagy is ubiquitin-independant
Autophagy in Cancer
Basal autophagy prevents DNA damage by degrading reactive oxidative species (ROS)
Causes accumulation of LC3, increasing phagophore formation
Cancer progresses > nutrient deprivation in tumour microenvironment promotes autophagy
Bortezomib (proteasome inhibitor) successful for treating multiple myeloma
prolonged inhibition activates autophagy which cancer cells exploit
Insoluble Protein Deposits (IPOD)
Centre of highly insoluble protein aggregates
Spatially distinct from JUNQ, multiple can exist in a cell
Proteins localised to IPODs are terminal
ATG8 proteins are highly associated
Hsp40 and 70 also localised
Similar to JUNQ mother cell also retains the deposit