DNA organization , segregation & DNA recombination
1/54
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
55 Terms
End-to-end length of Human DNA
1.02 m
Human DNA needs to be organized in less than 2000 micrometers cubed of space
DNA Packaging/Organization
Involves several levels
supercoiling
The following mechanisms are required to efficiently and effectively organize the DNA into a usable form:
Nucleosomes
Chromatosomes
Solenoids
looping onto matrix
The following mechanism is required to produce a mitotic chromosome
Higher order coiling
Proteins that organize chromosomes are essential and provide a mechanism for condensation, segregation, and organization of chromosomes
Virus
A non-cellular infectious particle genome containing a small nucleic acid genome with a limited number of genes
The nucleic acid can be single-stranded or double-stranded
The nucleic acid can be DNA or RNA
Physical size and genomic size varies a lot
Bacteriophage
A virus that infects (“eats”) bacteria
Capsid
Protein coat that contains viral genetic material
Non-enveloped virus
Contains genetic material in only a protein shell
Enveloped virus
Has an envelope of host cell cytoplasmic membrane surrounding the capsid
Bacteria
Haploid genomes
Most often have a single dsDNA chromosome
Some have more than one chromosome
Nucleoid
Bacterial chromosomes are densely packed to form a small region called the ____
Organized into a series of tight loops
Allow for efficient packaging of relatively long DNA molecules into very small spaces
Bacterial chromosome compaction
Proteins help organize the DNA into loops that pack the chromosome into the nucleoid
The circular DNA undergoes supercoiling
Small nucleoid-associated proteins
Participate in the DNA bending that contributes to folding and condensation of bacterial chromosomes
Structural maintenance of chromosome (SMC) proteins
Holds DNA in coils or V-shapes, maintains the shape facilitated by small nucleoid assisted proteins
Supercoiling
Covalently closed circular chromosomes exist in various ___ forms
The relaxed circle is the least twisted
_____ compacts DNA as a result of over or under rotations of helical twisting
Chromatin
The DNA and associated proteins of eukaryotic chromosomes
Chromosomes
Composed of half DNA and half protein
Histone proteins
Makeup half of the proteins in chromosomes, small basic proteins that tightly binds DNA
5 types in chromatin:
H1 - Linker DNA
H2A - Nucleosome
H2B - Nucleosome
H3 - Nucleosome
H4 - Nucleosome
Tightly conserved among eukaryotes
The remaining half of proteins, non-____ proteins, are very diverse and perform a variety of tasks in the nucleus
Nucleosome
Most basic unit of DNA packaginf
8 histone proteins within the core
2 of each: H2A, H2B, H3, and H4
Formed by a span of DNA that is wound around each protein octamer 1.65 times (~146 bp)
In humans, there is ~50 bp space between core nucleosomes (linker region)
Nucleosome assembly
Histones H2A and H2B assemble into H2A-H2B dimers, histones H3 and H4 assemble into H3-H4 dimers
Two H3-H4 dimers form a tetramer, then two H2A-H2B dimers associate with it to form the octamer
First level of DNA condensation
The wrapping of DNA around the octamer - compacts the DNA about sevenfold. E.g. 7m → 1m
Chemical modification
The ends of the histone proteins lie outside of the nucleosome and can be ___ _____
E.g.
Methylation
acetylation
phosphorylation
All associated with regulating gene expression
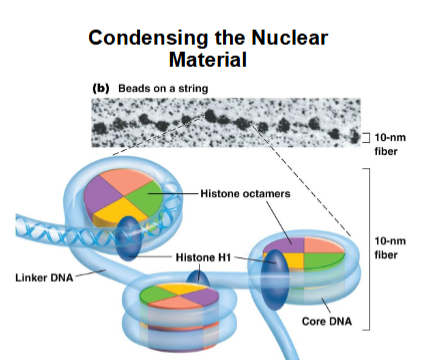
Electron micrographs of chromatin
In its least condensed state, it show a 10-nm fiber, or “beads-on-a-string” morphology-the “beads” are the nucleosomes
Kornberg
Proposed the nucleosome-based model of chromatin in 1974
The variable-length “string” between nucleosomes is linker DNA
H1 may associate with linker DNA
Chromatosome
H1 with a nucleosome may sometimes be called a ____
Histone H1 binds to DNA at edges of histone particle
covers another ~20 bp of DNA (total now ~166 bp) from 146 bp
DNA now wrapped about 2x around the histones
Solenoid structure
The 10-nm fiber is not observed under normal cellular conditions
a 30-nm fiber (x6 more condensed) is observed and forms when the 10-nm fiber coils into a ____ ___, with six to eight nucleosomes per turn and histone H1 stabilizing the solenoid
______ chromatin is lopped and attached periodically to a nonhistone protein chromosome scaffold
The loops on the scaffold form the 300-nm fiber
This is the state of the chromatin in a functioning cell (interphase)
Matrix attachment regions (MARs)
Chromatin loops of 20-100 kb are anchored to the chromosome scaffold by non-histone proteins at sites
Radial loop-scaffold model
Suggests that the loops gather into “rosettes” and are further compressed by non-histone proteins
Metaphase chromatin is compacted 250-fold compared to the 300-nm fiber
Euchromatin
Regions that contain actively expressed genes are less condensed during interphase
Open structure, DNA is accesible to enzymes (solenoidal form of chromatin) - expressed genes are found here
Heterochromatin
Regions that remain condensed in interphase and contain many fewer expressed genes
Often associated with the methylation of histones
Compact, inactive DNA common at centromeres and telomeres (no genes)
Comes in two forms:
Facultative
Constitutive
Facultative heterochromatin
Not always heterochromatin E.g. the sex chromosomes
Exhibits variable levels of condensation, related to levels of transcription of resident genes
Inactivation is reversible
Constitutive heterochromatin
Permanently condensed
Found prominently in centromeres and telomeres
composed primarily of repetitive DNA sequences
Dynamic chromatin structure
Changes in level of compaction regulate access to DNA by proteins for replication, transcription, recombination, or repair
E.g. chromosome centromeres have constitutive heterochromatin
But when they replicate in S phase, heterochromatin dissipate
Nucleosome core particles dissociate from DNA ahead of replication fork and re-form centromeric heterochromatin after the fork passes
Variable borders for reestablishing centromeres and typically no impact on gene expression
Gene expression
Can be controlled by the state of chromatin in which a gene is located
Can be dictated by chromatin structure, which is transmissible from one cell generation to the next
Hermann Muller
The eyes of the fruit fly Drosophila are red, thanks to the expression of the w locus that is usually in a region of euchromatin near the telomere of the X chromosome
He used X-rays and isolated mutants in which a segment of the X chromosome had inverted thereby placing the w locus near the centromere
The result was that in some eye cells, the gene was not expressed due to the spread of heterochromatin to the centromere; while other cells it was expressed giving a variegated appearance to the eyes
Germ line cells
Sperm and ovum
Chromosome segregation
The process in eukaryotes by which two sister chromatids formed as a consequence of DNA replication (or paired homologous chromosomes) separate from each other and migrate to opposite poles of the nucleus
Occurs during both mitosis and meiosis
Also occurs in prokaryotes, in contrast to eukaryotic ___ ___, replication and segregation are not temporally seperated but is simultaneous instead of being sequential like in eukaryotes. It is progressive after following replication
The cell cycle
1) Interphase
2) M phase or Mitosis
Interphase
Occurs between cell divisions, chromosome/DNA replication occurs
The nucleus is granular looking
DNA is replicated
Homologous chromosomes
Very similar to each other and have the same size and shape
They carry the same type of genetic information, so they have the same genes in the same locations
Prophase
Chromosomes condense
Nucleolus dissapears
Centrioles move to poles
spindles form
Nuclear membrane breaks down
Prometaphase:
Movement of chromosomes to centre of the cell
Metaphase
Chromosomes align at equatorial plane of the cell
Mitotic spindle formation complete
Anaphase
Starts when sister chromatids split
chromosomes move to poles of cell
cell begins to elongate
cleavage furrow starts
Telophase
Begins when chromosomes reach the poles
Nuclear membrane reforms (shady colour)
chromosomes de-condense
nucleoli reform
spindle fibers disappear
cleavage furrow continues
Cytokinesis
Not part of mitosis but normally follows closely after mitosis
Equals completion of cleavage furrow and production of two cells
Not always part of the M phase but this is not always true - Drosophila development
Meiosis
It is a specialized type of mitosis
Occurs in the germ line/cells
Necessary for the production of gametes in diploid, sexually reproducing organisms
Involves two sequential cell divisions without DNA replication between divisions
8 stages in total
Happens in germ cells
Purpose is sexual reproduction
Produces 4 haploid daughter cells
Chromosomes number is halved in each daughter cell
Genetic variation increased
Meiosis 1
Reduction division step
Segregation of homologous chromosomes reduction, diploid to haploid
Odd step
More complex
Subdivided into more steps
Leptotene
1st stage of prophase 1
Chromosomes begin to condense and become visible
Thickened regions (chromomeres) appear
Zygotene
2nd stage of prophase 1
Homologous pair (bivalent) of chromosomes
Chromosomes continue to condense and there is active pairing of the chromosome threads between non-sister chromatids
Prophase 1 - Pachytene
3rd stage of prophase 1
Chromosomes become fully aligned
Slow dissociation of nuclear envelope
Allows for recombination
Diplotene
4th stage of prophase 1
Aligned homologous pairs become less tightly aligned
Chiasmata appear and mark the locations where crossing over has occured
Diakinesis
5th stage of prophase 1
Compaction is completed and the chromosomes are ready to be segregated
Telophase 2
1 out of 4 recombinant from the mother
1 out of 4 recombinant from the father
The 2 others are non-recombinant
How meiosis differs from mitosis
Reduction in the number of chromosomes (from diploid to haploid)
recombination between chromosomes
Importance of meiosis
Recombination increases diversity even more by reshuffling genetic information between the chromosomes
The total is not the sum of the parts
Production of haploid cells by meiosis is a critical component of sexual reproduction
Independent assortment of chromosomes during meiosis 1 (Mendel’s fourth law) produces diversity in offspring (two offspring will almost never have exactly the same chromosome complement)
Mendel’s fourth law
Also known as Independent assortment
Occurs during meiosis 1, and produces diversity in offspring which ensures that they will almost never have exactly the same chromosome component
8 possible combinations with only 3 chromosomes
Mitosis
4 stages in total + interphase
Happens in somatic cells
Purpose is cell proliferation
Produces 2 diploid daughter cells
Chromosome number remains the same
Genetic variation doesn’t change