3.8 The control of gene expression
1/95
Earn XP
Description and Tags
- Alteration of the sequence of bases in DNA can alter the structure of proteins - Most of a cell’s DNA is not translated - Regulation of transcription and translation - Gene expression and cancer - Using genome projects - Recombinant DNA technology - Differences in DNA between individuals of the same species can be exploited for identification and diagnosis of heritable conditions - Genetic fingerprinting
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
96 Terms
start and stop codon
at the start of every gene there is a start codon, this codes codes for the amino acid (initates translation)
at the end of every gene there are 3 bases that do not code for an amino acid - marks the end of a polypeptide chain - stop translation
what are the 3 feature of genetic code ?
Degenerate -more than one code to code for each amino acid (64 combinations only need 20) remember the genetic code wheel
Overlapping - each base in a gene is only part of one triplet of bases that code for one amino acid
Universal - the same triplet of bases codes for the same amino acid in all organisms this is why the genetic code is described as universal
introns and exons
In most eukaryotic genes, coding regions (exons) are interrupted by noncoding regions (introns). During transcription, the entire gene is copied into a pre-mRNA, which includes exons and introns. During the process of RNA splicing, introns are removed and exons joined to form a contiguous coding sequence.
what are the 6 type of gene mutations ?
addition
deletion
inversion
substitution mutation
duplication
translocation
What is addition in mutations?
one extra base added to the sequence - all subsequent codons are altered - causes FRAME-SHIFT
All the altered codons could potentially form different amino acids - resulting in different sequence of amino acid - creating non-functioning protein
What is a deletion mutation?
one nucleotide is taken away from a gene or DNA sequence
this causes FRAME SHIFT
could result in different polypeptide chain and non functioning proteins
What is a substitution mutation?
one nucleotide takes place of another - however the number of bases remain the same - NO FRAME SHIFT - results in only one codon changing & due to genetic code being degenerate it may code for the same amino acid- therefore no impact AKA SILENT MUTATION (wheel)
triplet/codon codes for different amino acid
What is inversion mutation?
A section of bases that detach from the DNA sequence,but when they re-join they are inverted, so the section of code is back to front- this results in different amino acids being coded for in this region.
What is translocation ?
A section of bases on the chromosome detaches and attaches onto a different chromosome
this is a substantial alteration and causes significant impact on the gene expression and therefore the resulting phenotype
What are the key causes of mutation ?
- randomly and spontaneously during DNA replication - interphase cell cycle
- mutagenic agents
- ionization radiation
What are mutagenic agents?
Factors that increase the rate of gene mutation
increases the frequency of spontaneous mutations if exposed to mutagenic agents
High energy & ionization radiation
alpha and beta , x-rays and gamma rays -cause damage & disrupt the structure of DNA
Carcinogens - alter the structure of DNA & interfere with transcription ( copy error from mRNA )
How mutations occur
Alterations to one or more nucleotide bases in the sequence of DNA for one gene.
occurs DURING DNA REPLICATION DURING INTERPHASE IN CELL CYCLE
when the bases change/mutate in DNA the triplet codon changes bases meaning it changes the mRNA complementary pair (from the original vs mutated )
meaning this codes for a different amino acid sequence in the encoded polypeptide to make protein
if sequence of amino acids changes then when protein is modified into the tertiary structure it will form hydrogen & ionic bonds in a different 3D shape - creating a non functioning protein
most mutations do not alter polypeptide if do its only slightly (silent mutation) bc genetic code is degenerate
DNA - mitochondria + chloroplast endosymbiotic theory
the presence of DNA in mitochondria and chloroplast is explained by this theory,
it states that bacterial cells were engulfed by larger cell during evolution
- the bacteria and host cell formed a beneficial symbiotic relationship
- the bacterial cell becomes incorprated into the larger cell to become organelles(host cell)
when chloroplast and mitochondria were free living their own needed DNA to survive - this explains why chloroplasts and mitochondria still have some DNA
DNA chloroplasts encodes for photosynthesis
DNA mitochondria encodes for respiration
define stem cells
Stem cells are undifferentiated cells that can continually divide and become specialised. Differentiation is the process by which stem cells become specialised
Name the different types of stem cells that have different differentiation abilities
Totipotent
pluripotent
multipotent
unipotent
extra information :
Any totipotent stem cell is capable of forming a complete viable offspring
Pluripotent stem cells are slightly differentiated cells. They can give rise to nearly all the cell types but are not able to form a complete viable offspring
Multipotent stem cells are somewhat more differentiated cells. They can give rise only to cells of closely related families.
What are totipotent stem cells
These stem cells can divide and produce any type of body cell(can mature into any body cell). During development totipotent cells translate only part of their DNA resulting in cell specialisation(during the process of cell specialisation, only some of the genes are expressed. This means that only part of the DNA of a cell is translated into proteins. The cell therefore only makes those proteins that it requires to carry out its specialised function.)
Totipotent cells occur only for a limited time in early mammalian embryos and because of this it is not used in as much research as pluripotent because it is less available after cell division in comparison of pluripotent

What are pluripotent stem cells and its source
these stem cells are found in embryos and can divide in unlimited numbers and differentiate into most types of cell.
using them in research to treat human disorders. these stem cells could be used to regrow damaged cells in human such as replacing burnt skin cells or beta cells for type 1 diabetes.
there are issues with this as sometimes the treatment doesn’t work or the stem cells continually divide to create tumors
additional ethically there is debate on whether it is right to make therapeutic clone of yourself to make embryo to the stem cells to cure a disease and then destroy the embryo
[ embryos up to 16 days after fertilisation contain pluripotent stem cells and can differentiate into any type of cells ] - source of stem cell
what are multipotent stem cells and its sources
found in mature mammals and can divide to form a limited number of different cell types e.g. bone marrow can differentiate into a limited number of cells.
[ umbilical cord blood contains multipotent adult stem cells + the placenta has multipotent stem cells and can develop into a limited number of specialized cells] - sources of stem cell
order the stems from most to least differentiated
1) Unipotent stem cells are the most differentiated(specific) cells can give rise to only a particular cell type to which they belong also known as adult stem cells
2) Multipotent stem cells are a bit less differentiated cells. They can give rise only to cells of closely related families. also known as adult stem cells
3) Pluripotent stem cells are slightly differentiated cells. They can give rise to nearly all the cell types
4) Totipotent stem cells are the actual stem cells. They are completely undifferentiated but can differentiate/become specialised into any specific cell type. also known as embryonic stem cells
what are unipotent stem cells
these are found in mature mammals and can be divided . Unipotent cells can only differentiate into one type of cell (differentiate into the same cell)
I’m assuming it has less ethical issues because
why do unipotent stem cells have fewer ethical issues?
Unipotent stem cells have less ethical issues because they are derived from adult tissues, not embryos. they can only develop into a specific type of cell. This eliminates concerns about destroying embryos and avoids the ethical debate surrounding embryonic stem cells.
Stem cells originate from various sources in mammals:
Embryonic stem cells
Umbilical cord blood stem cells
Placenta stem cells
Adult stem cells (bone marrow) from persons own body
Induced Pluripotent Stem Cells
iPS cells created from adults unipotent cells using appropriate protein transcription factors to overcome some of the ethical issues with using embryonic stem cells.(pluripotent found in embryos)
iPS cells are created from adult unipotent cells. These cells which can be from most body cells, are altered in the lab to return them to a state of pluripotency.- more variation/diversity differentiates into different types of cells than unipotent that at only divide via mitosis to form the same type of cell
To do this the genes that were switched off to make the cell specialized(unipotent) must be switched back on this is done using transcriptional factors.
They are very similar to embryonic pluripotent stem cells but do not cause the destruction of an embryo and the adult can give permission (for example, can be taken from a donor's bone marrow, and from a person's circulating blood.) - so requires permission from adult whereas embryonic stem cells do not get permission from embryo
the iPS has shown a self-renewal property in that they can divide indefinitely to give limitless supplies. for these reasons, they could be used in medical treatment instead of embryonic stem cell
What is iPS and its advantages ?
Induced pluripotent stem cells are unipotent stem cells that have been reprogrammed to become pluripotent by using protein transcription factors to express genes associated with pluripotency.
ADVANTAGES
1) doesn’t cause embryonic destruction
2) it has self-renewal property so can divide indefinitely to give a limitless supply
3) its used in medical treatment instead of embryonic stem cells removing ethical issues
4) no ethical issues no cloning or embryo destruction involved
what are the different ways that control gene expression
epigenetic control
role of transcription factors
oestrogen in regulating transcription
RNA interference(RNAi)
define epigenetics
Epigenetics involves heritable changes in gene function(preventing gene expression), without changes to the base sequence of DNA. controls gene expression
these changes are caused by changes in the environment and can inhibit transcription
define epigenome
chemical tags that affect the shape of the DNA-histone complex
changes in the environment and its impact on the epigenome
factors such as diet stress and toxins can add epigenetic (chemical tags) to the DNA and this can control gene expression in eukaryotes.
epigenetic control is mediated by chemical tags the epigenome impacts the shape of the DNA-histone complex and whether the DNA is tightly wound to histone so won’t be expressed or unwound so it will be expressed
if the DNA is tightly wound around the histone protein then transcription factors cannot bind. Therefore the epigenome which is due to changes in the environment can inhibit transcription.
what are the 2 key chemical tags that affect the shape of the DNA-histone complex ?
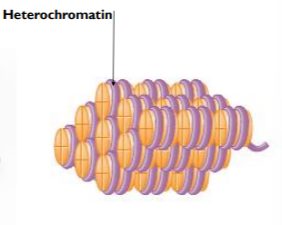
METHTLATION OF DNA - results in tightly wound - heterochromatin
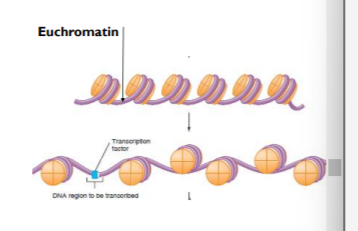
ACETYLATION OF HISTONE PROTEINS - results in loose wound - euchromatin
Acetylation _________ rate of transcription whereas methylation _____________ the rate of transcription
increase
decrease
heterochromatin
when DNA is tightly wound around the histone proteins the genes are inactive/off because their promoter regions are inaccessible to transcription factors
METHYLATION CAUSES DNA TO BECOME MORE CONDENSED THEREFORE DNA LESS ACCESSIBLE TO TRANSCRIPTION FACTORS
*condense in this context means to be more dense/tightly packed
Euchromatin
when DNA is less condensed around the histone proteins the genes can be turned on/activated. this is because their promoted regions are now accessible to transcription factors
ACETYLATED CAUSED DNA TO BECOME LESS CONDENSED - THEREFORE LESS TIGHTLY WRAPPED MAKING DNA MORE ACCESSIBLE TO TRANSCRIPTION FACTORS
HOWEVER, THE SPEC FOCUSSES ON THE DECREASED ACETYLATION OF HISTONES WHICH CAUSES CHROMATIN TO CONDENSE SO TRANSCRIPTION FACTORS + RNA POLYMERASE CAN’T BIND
*condense in this context means to be more dense/tightly packed
1) euchromatin ____________ methylation and _______________ acetylation of associated histone proteins
2) heterochromatin ________ methylation and __________ acetylation of associated histone proteins
1) decrease + increase
2) increase - decrease
INCREASED Methylation of DNA (cpG) - note it affects dna
add on a methyl group, these methyl groups can only attach to DNA specifically cytosine base
increased methylation of DNA inhibits transcription because it packs tightly together and this prevents transcription factors from binding to promoter regions of DNA and prevents a section of DNA from being transcribed
methyl = +ive charge
DNA = -ive charge
causes DNA and methyl to attract and tightly coil together
the attachment of methyl also attracts proteins that (more) condense chromatin so preventing transcription as RNA polymerase can’t bind.
so gene is turned off/inactive
DECREASED Acetylation of histones - note it affects histones
acetyl group = -ive charge
acetyl groups are removed from histones so histones become more positively charged and this attracts phosphate groups on DNA as phosphate is negatively charged
this makes the DNA and histones more strongly associated and hard for the transcription factors to bind.- inhibits transcription
treatment of disease- tumours
increased methylation/decreased acetylation= decrease in expression of tumour suppressor genes so inhibition of tumour suppressor genes (tumour formation) + proto-oncogenes(which inhibits tumour formation)
decreased methylation/increased acetylation = increase in expression of tumour suppressor genes, so tumors are less likely to form
for context, this links to flashcards above e.g. increased methylation inhibits binding of transcription factors because it is too tightly packed so the gene would turn off/inactive so the tumour suppressor gene wouldn’t bind so tumours are more likely to form.
e.g unipotent cell genes are off because its specialized therefore is more condensed so gene is switched off to alter it to a pluripotent state we turn on these genes by making chromatin less condensed by increased acetylation allowing it to bind to transcription factors.

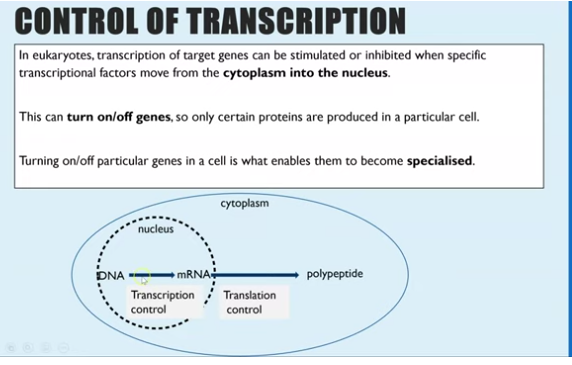
what controls transcription in eukaryotes
transcription of target gene can be either inhibited or stimulated when a specific transcription factor moves from the cytoplasm tot he nucleus
controlling gene expression (turning “off“ or “on”) makes cells specialised
transcription factors are _________ that move from the ____________ to the nucleus
proteins/molecules
cytoplasm
how do transcriptional factors affect the control of gene expression ?
transcription of a gene will only occur when the transcription factor from the cytoplasm enters the nucleus via the nuclear pore where it binds to DNA in the promoter/operator regions in the nucleus.
binding to promoter - initiates transcription and therefore initiates translation of the protein
binding to operator - inhibits transcription and therefore inhibits translation of gene
these protein molecules are called transcription factors and each one can bind to a different base sequence on DNA which enables RNA polymerase to bind therefore initiate transcription genes .
transcription begins creating mRNA molecules for that gene which can then be translated in the cytoplasm to create the protein. without the binding of a transcription factors the gene is turned off so the protein isn’t made (operator)
what does it mean by promoter and operator regions in dna ?
binding to promoter - initiates transcription RNA polymerase can attach to the promoter and therefore initiates translation of the protein
binding to operator - inhibits transcription and therefore inhibits translation of gene
the role __________ have in initiating/activating transcription factors
Oestrogen
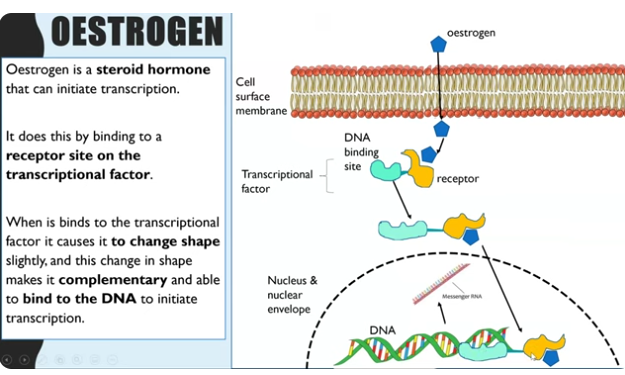
OESTROGENS role in initiating transcription by transcription factors ?
oestrogen is a steroid hormone - meaning it is lipid soluble (outside of a cell oestrogen is transported with blood and then diffuse through cell membrane) entering the cell
once in the cell it enters cytoplasm and binds to the receptor site on transcription factor because oestrogen is complimentary in shape to the receptor. (oestrogen receptor)
change in the tertiary structure of the DNA binding site so the transcriptional factor is now complimentary to bind to DNA promoter region
This activates the transcriptional factors to be able to move into the nucleus bind to the promoter region which stimulates RNA polymerase increasing transcription.
(remember transcription of a gene can only occur if transcription factors bind to the DNA)
transcription factors have two sites - receptor site and DNA binding site
RNA interference RNAi - what are the two types?
siRNA
miRNA
RNAi - process of regulation transcription
translation of mRNA of target genes can be inhibited by RNAi mRNA this is when an mRNA molecules that has already been transcribed gets destroyed before translation to form polypeptide chain and this is done by small interference RNA (siRNA )
how siRNA works to prevent translation ?
an enzyme can cut mRNA stranded ) into siRNA (double stranded)
One strand of the siRNA then combines with another enzyme
The siRNA - enzyme complex will then bind via complementary base pairings on one specific type of mRNA molecule (becomes double stranded)
cells cant recognise double stranded mRNA so the enzyme will cut up/degrade the mRNA so it cannot be translated.
miRNA acts similar but is less specific so can bind to many mRNA types
benign tumours characteristics
these can grow very large but a slow rate
non-cancerous because they produce adhesive molecules sticking them together and to a particular tissue
often surrounded by a capsule so they remain compact and can be removed by surgery and rarely return - doesn’t spread
the impact if localised
often not life threatening depends on the location
malignant tumour characteristics
these are cancerous and grow large
the cell nucleus becomes large and the cell can become specialised again
they do not produce the adhesive so instead metastasise, meaning the tumour breaks off and spreads to other tissues in the body
the tumour is not encapsulated and instead can grow projections into surrounding tissues and develop its own blood supply
it can be life threatening and the removal of the tumour needs supplementary treatment (radiotherapy/chemotherapy) and recurrence is more likely
tumour development
tumour develop due to a gene mutation in either the tumour suppressor gene and/or oncogene, the abnormal methylation of tumour suppressor gene and oncogenes or increased oestrogen concentration
what is a proto-oncogene
Proto-oncogene are normal genes that code for proteins that regulate cell growth (growth factors) and cell differentiation
stimulate cells to divide by producing proteins that stimulate mitosis cell division when the body needs new cells
what is an oncogene ?
Oncogenes are mutated genes that have the ability to cause cancer through the deregulation of cell growth
encodes proteins involved in DNA replication and mitosis and can result in this process being permanently switched on/activated to make cells divide continually + tumour formation
hypomethylation(less) -turn on
hypermethylation (more)- turn off
tumour suppressor genes causing cancer
these genes produce/encodes proteins to slow down cell division and to cause cell death if DNA copying errors are detected
if a mutation occurs in the tumour suppressor gene not producing proteins to slow down cell division then cell division could continue and mutated cells would not be identified and destroyed .leading to uncontrollable cell division and tumour formation.
hypermethylation leads to increased expressions of tumour formation because chromatin is more condensed
abnormal methylation to cause cancer
methylation can cause a gene to turn off or on
The tumour suppressor gene could become hypermethylated, meaning an increased number of methyl groups attached to it. this results in the gene being inactivated and becomes turned off - proteins wont be produced to slow down cell division - forming tumour
the opposite could occur in oncogene - increases expression of oncogenes as they may be hypomethylated reducing the number of methyl groups attached. this results in the gene being permanently switched on - lots of proteins produced to initiate cell division constantly- forming tumour
increased oestrogen concentrations to cause cancer
oestrogen is produced by the ovaries to regulate the menstrual cycle, but after the menopause this stops
instead fat cells n the breast tissues can produce oestrogen and this had been linked with causing breast cancer in the women post-menopause s oestrogen activated transcription factors and consequently the transcription of genes e.g. oncogenes
this has a knock on effect as the formation of tumour results in even more oestrogen production (as tumours rely on the hormone to increase division and therefore survive) which increase the tumour size and attracts white blood cells which can increase the tumour size further.
this could be because oestrogen can activate a gene by binding to a gene that initiates transcription and if this is a proto-oncogene the result is permanently turned on and activating cell division
genome and what are sequencing projects ?
is the entire genetic material of an organism in the nucleus of a cell in eukaryotes
sequencing a genome means working out the DNA base sequence for all the DNA in a cell.
read the genome of a variety of organisms
determining genome of simple(no introns) organisms allows the sequence of the protein that are encoded
application including identification of potential antigens for use in vaccine production
Sequencing methods are continuously updated and have become automated.
what is a proteome ?
the complete set of proteins expressed by an organism can be eoncoded by the genome
simpler organisms vs complex organisms
hints: introns
simple organisms like prokaryotic cells do not contain introns in their DNA. this means that the genome can be used directly to sequence the proteins that derive the genetic code of the organisms
this is very useful for many reasons including identifying potential antigens to use in a vaccine
more complex organisms eukaryotes have introns and regulatory genes in their DNA due to this the genome cannot easily be used to translate the proteome
what are the three recombinant DNA technologies
creating DNA fragments
genetic fingerprinting
Genetic screening,counselling and locating genes
what does transgenic mean and why can the same proteins code for the same thing to all organisms ?
as the genetic code is universal, genes from one organism will code for the same proteins in all organisms
transgenic organisms have been genetically modifies
their DNA is recombiant - combined from two different organisms
recombinant DNA technology
the combining of different organisms DNA which could enable scientists to manipulate and alter genes to improve industrial process and medical treatment
Recombinant DNA technology involves the transfer of fragments of DNA from one organism, or species, to another. Since the genetic code is universal, as are transcription and translation mechanisms, the transferred DNA can be translated within cells of the recipient (transgenic) organism.
the first step in this technology is to produce or isolate the fragments of DNA to be recombined with another piece of DNA
there are three methods to create fragments of DNA : forming or isolate DNA fragments
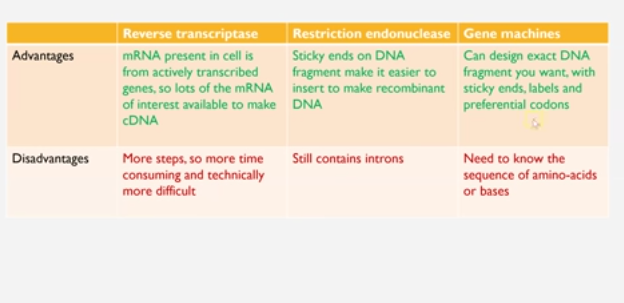
1) reverse transcription
2) restriction endonuclease
3) gene machine
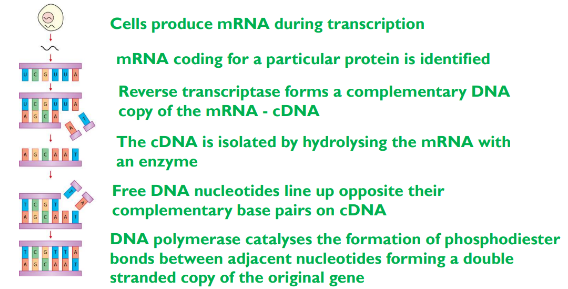
1) Reverse transcriptase to create fragments - conversion of mRNA to complementary DNA (cDNA)
process
the enzyme makes DNA copies from mRNA
naturally occurs in retroviruses (RNA as main genetic material not DNA ) e.g HIV
the cell that produces the protein that scientists want is chosen and should have a large amount of mRNA for the protein
the reverse transcriptase enzyme aligns and joins the complementary DNA bases with the mRNA bases
this is single stranded and is called complementary DNA (cDNA)
to makes this DNA fragment double stranded the enzyme DNA polymerase makes the cDNA double stranded (joins adjacent nucleotide to form phosphodiester bonds)
the cDNA doesn’t have introns which is and advantage
restriction endonucleases - using restriction enzymes to cut a fragment containing the desired gene from DNA
restriction endonucleases are enzymes that cut up DNA at specific restriction sequence to form DNA fragements
there are many restriction enzymes/endonucleases that have an active site complementary in shape to a range of different DNA base sequences described as recognition sequences and therefore each enzyme cuts the DNA at a a specific location by breaking phophodiester bond between adjacent nucleotides
some enzymes cut at the same location in the double strand DNA and create a blunt end other enzymes cut to create staggered ends with exposed DNA bases - overhanging bases
the exposed staggered ends are palindromic (same but reads backwards )and referred to as sticky ends because they have the ability to join DNA with complementary base ends and are helpful in cloning because they hold two pieces of DNA together so they can be linked
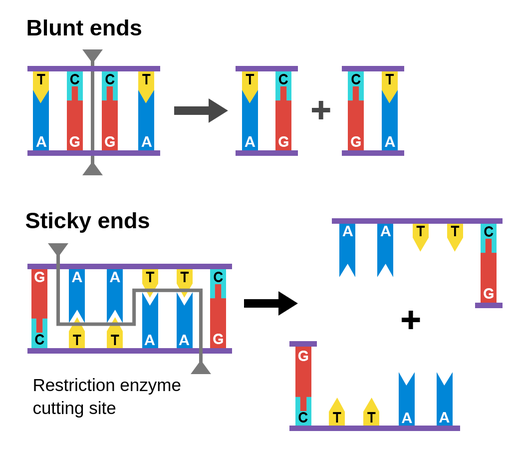
explain how sticky ends are useful in genetic engineering ?
joins two pieces of DNA by complementary base pairing
gene machine - creating the gene in a ‘gene machine’.
DNA fragments can be created in a lab using computerised machine
scientists first examine the protein of interest to identify the amino acid sequence and from that work out what the mRNA and DNA sequence
The DNA sequence is entered into the computer which checks for bio safety and bio security that the DNA being created is safe and ethical to produce
the computer can create a small section of overlapping single strands of nucleotide that make up the gene called oligonucelotides
the oligonucleootides can be joined to create the DNA for the entire gene
PCR (polymerase chain reaction) can be used to amplify the quantity and to make the double strands
this process is very quick, accurate and makes introns free DNA so can be transcribed in prokaryotic cells
advantages and disadvantages of all three of the methods to create fragments of DNA
fragments cloned to amplify sample
in vitro: PCR (polymerase chain reaction)
in vivo
in vivo cloning - cloned within living organims
produce DNA fragments of genes of interest
insert DNS fragment into a vector
transform a host cell with the vector
identify transformed cells
grow the host cell (clone/make copies of the gene)
1.promoter and terminator region
restriction endonucleases enzyme used to cut out the gene - DNA fragments of interest DNA fragments must be modified to ensure transcription of this gene can occur
promoter regions must be added at the start of the DNA fragments. This is a sequence of DNA which is the binding site for RNA polymerase to enable transcription to occur
a terminator region must be added at the end of the gene. it causes RNA polymerase to detach and stop transcription, so only one gene at a time is copied into mRNA
2.insert DNA into a vector
vector can carry the isolated DNA fragment into the host cell, plasmids are the most common vectors - circular DNA, separates from main bacterial genome which only contains a few genes
the plasmids is cut open using same restriction endonucelease. this creates the same sticky ends. therefore the DNA fragment sticky ends (exposed nucleotides) are complementary to the sticky ends on the plasmids
The DNA fragment and cut plasmid are combined and the enzyme ligase sticks them together (anneals them) Ligase catalyses the condensation reaction to form phosphodiester bonds between nucleotides
transform a host cell with the vector/transformation
the vector (plasmid with the recombinant DNA) next needs to be inserted into the host cell, where the gene will be expressed to create the protein required
to do this the cell membrane of the host cell must be more permeable. to increases the permeability, the host cells are mixed with Ca 2+ and heat shocked (sudden increase in temperature
this enables the vector to enter the host cells cytoplasm
identity transformed cells why?
not all the host cells (usually bacteria) will successfully take up the recombinant plasmid why?
3 issues can occur :
the recombinant plasmid doesn’t get inside the cell
the plasmid re-joins before the DNA fragment entered
the DNA fragment sticks to itself, rather than inserting into the plasmid
That is why scientists must identify which bacterial successfully took up a recombinant plasmid
Marker genes - on the plasmids can be used to identify which bacteria successfully took up the recombinant plasmid.
Three different marker genes used are:
antibiotic resistance genes
genes coding for fluorescent proteins
genes coding for enzymes
antibiotic resistance marker genes
insert a gene that makes bacteria resistant to tetracyclines gene and resistant to ampicillin next insert DNA fragments to be inserted
resistance to tetracycline gene disrupt by DNA fragment, this gene will no longer create a functional protein - so any bacteria that contain that recombinant plasmid will not be resistant to that antibiotic tetracycline
next, we grow bacteria on agar, we work out from the bacteria which of then successfully take up the recombinant plasmid. we use a sterile velvet block and place this sheet and stamp in on a petri dish with ampicillin antibiotic in the agar
any of the colonies that still grow tell us they must have the plasmid in to having the gene that makes it resistant so can grow even in ampicillin antibiotic
lasts step we then use sterile velvet block to take imprint on a perti dish wish as tetracycline antibiotic dissolved with agar we can see what colonies still grow
"https://www.youtube.com/embed/FzepP22pvww?list=PLOfYYgIrtVMhkt6bDzeTdTkEm5l6RkSfa"
fluorescent markers
green fluorescent protein GFP. Can be inserted into the bacteria plasmid.
DNA fragment is inserted in the middle of the GFP gene this disrupts it and prevents GFP production. those that contain recombinant don’t produce green glowing
enzyme markers
the enzyme lactase can turn a certain substance blue from colorless. the gene for this enzyme is inserted into the plasmid. then a DNA fragment is inserted in the middle of the lactase gene. This disrupts it and prevents lactase production. the bacteria are then grown on an agar plate with the colorless substance. the colonies that cannot turn colorless substance blue contain the recombinant plasmid
grow the host cells (clone/make copies of the gene)
a fermenter is used to grow multiple copies of the host cell identified as containing the recombinant plasmid. This large cloned population of the host cell can produce the protein coded for by the inserted DNA fragments (e.g bacteria producing insulin from the inserted insulin gene)
in vitro cloning - not within a living thing
PCR
amplifying DNA fragments once the DNA fragments have been isolated, they needs to be cloned to create large quantities, this can be done by invivo or invitro
in vitro- fragments of DNA cab be amplified in vitro via the polymerase chain reaction (PCR). This is done an automated machine
equipments :
thermocyler
DNA fragment to be amplified
DNA polymerase - taq polymerase
primers- short strands of single-strand DNA
DNA nucelotides
PCR method
the temperature is first increased to 95 degrees to break hydrogen bonds and split the DNA into single strands (denaturing)
the temperature is then decreased to 55 degrees so that the primer can attach (annealing)
the enzyme DNA polymerase then attaches complementary free nucleotides and makes a new strand to align next to each template (synthesis). The temperature is increased to 72 degrees for this stage the optimum for taq DNA polymerase
advantages of PCR :
automated machine - more efficient
rapid - 100 billion copies of DNA can be made within hours
doesn’t require living cells - quicker and less complex techniques needed compared to invivo
DNA probe
short single-stranded DNA molecule that is designed to be complementary for a sequence to be detected. DNA probes are made in smaller quantities and then amplified using PCR. The DNA labeling of the fragments either uses radioactive isotopes or a fluorescent dye which glows under certain wavelengths of light
DNA probes can be used in order to detect heritable conditions of health risks.
Genetic fingerprinting - VNTRS
VNTRS - 95 percent of human DNA is made up of introns which consists of many Variable number tandem repeats.
the probability of 2 individuals having the same VNTRs is very low however the more closely related you are the more similar the VNTRs are.
Genetic fingerprinting is the analysis of VNTR DNA fragments and this can used to determine genetic relationships and the genetic variability within a population
6 processes of genetic fingerprinting1
collection
extraction
digestion
separation
hybridisation
development
analysis7
1. collection and extraction
the smallest sample of DNA can be collected for genetic fingerprinting. This could be from blood, body cells, or hair follicles. If the sample is DNA and is small then PCR is used to amplify the amount of DNA
2. digestion
we have larger sample of DNA that we want to examine by using restriction endonucleases - enzyme are added to cut the DNA into smaller fragments which’s active site is complementary tot eh sequence before VNTRS Enzymes which cut close to the target VNTRS are added
https://www.youtube.com/embed/ws25XREjnAA?list=PLOfYYgIrtVMhkt6bDzeTdTkEm5l6RkSfa"
separation
The DNA samples are loaded into small wells in agar gel. The gel is placed in a buffer liquid with an electrical voltage applied. DNA is negatively charged, so the DNA samples move through the gel towards the positive end of the gel
This stage is GEL ELECTROPHORESIS
the agar gel creates resistance for moving DNA and smaller pieces of DNA can move faster and further along the gel.
This is how the different lengths of DNA (VNTRs) are separated
an alkaline is then added to separate the double strands of DNA
Hybridisation
DNA PROBES are short single-stranded pieces of DNA complementary in base sequence to the VNTRs. The probes are radioactively or fluorescently labelled. DIFFERENT DNA probes are mixed with the single-stranded DNA VNTR’s on the agar gel for them to bind (hybridize)
Those that are complementary to VNTRS (single-stranded) bind
development
the agar gel will shrink and crack as it dries and therefore the VNTRs and DNA probes are transferred to a nylon sheet.
The nylon sheet can then be exposed to x-rays to visualise the position of radioactive gene probes, or UV light if fluorescence probes were used
Analysis
The position of the DNA bands are compared to identify genetic relationships. the presences of a disease causing gene and to match unknown samples from crime scenes
can be used in paternity tests
interpreting data showing the results of gel electrophoresis
compare the band of VNTRs
in the mother, child and the potential fathers, all of the child VNTRs must have been inherited from either the mother or father
USES OF GENETIC FINGER PRINTIING
Forensic science to place suspects are crime scenes
for medical diagnosis
to ensure animals and plants are not closely related b4 being breed - reduced passing on genetic conditions
DN probes
shory, single-stranded pieces of DNA that are labelled radioactively or fluorescently so they can be identified.
this is used to locate specific alleles of genes and to screen parents for heritable conditions. drug responses or health risks
DNA probes are created to have complementary base sequences to the allele that is being screened for
the patient’s DNA sample is treated to make it single-stranded and it it then mixed with the DNA probes
if the patient has the allele then the DNA probe will hybridize and label indicates the presence
DNA hydridisation
the patient’s DNA Sample is heated to make it single-stranded. the heat causes the hydrogen bonds between bases to break (denaturing)
the patients single-stranded DNA sample is mixed with the DNA Probe and cooled to and any complementary sequences can align and form hydrogen bonds (anneal)
Some of the patient DNA samples will anneal back together but some will anneal with the DNA probe.
method locating specific alleles of genes
to locate a specific allele, the DNA base sequence must be known to then create the DNA probe
this can be determined using DNA sequencing techniques
the fragment of DNA can be produced using a gene machine
this fragment can be amplified using PCR
the label is then added with radioactive nucleotide containing isotope 32 P or a fluorescent label which emits light under UV LIGHT
After hybridisation, the DNA is washed so that any unbound DNA probes are washed away
the presence of the radioactive or fluorescence label therefore indicates that the allele of interest is present in the patient’s DNA
genetic screening
this method can be used to screen for potential genetic disorders or the presence of cancer-causing oncogenes
it is possible to screen for multiple diseases simultaneously using an array where multiple different DNA probes are attached
why is genetic screening important
personalised medicine
this is one key reason for having your DNA screened can be advantageous
certain drugs such as painkillers are more or less effective depending on your genotype. it can also help determine the best dose, which increases the effectiveness, and safety and can save money
vitamin E can be given to diabetics to reduce the risk of CVD for some, but for others with a different genotype it can increase the risk
Genetic counselling
having your DNA screened is a decision that must be thought carefully and must be discussed with patients so they make an informed decision.
genetic counseling is like a type of social work where people can have their family history researched to consider the likelihood of them carrying any alleles linked to diseases before starting a family or for their general health. patients are informed of any potential risks to themselves or future offspring if they were to carry the allele
some examples include screening for cystic fibrosis and sickle cell anemia prior to starting a family. if there is a family history of breast cancer you may want to be screened for alleles linked to this cancer. if you find the allele is present oy can be screened few tumors more frequently, reduce environmental risk factors, or even opt for a mastectomy