lec 7.5 - RNA splicing
1/28
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
29 Terms
why do eukaryotes have introns? advantages?
alternative splicing of them allows for more proteins to be made
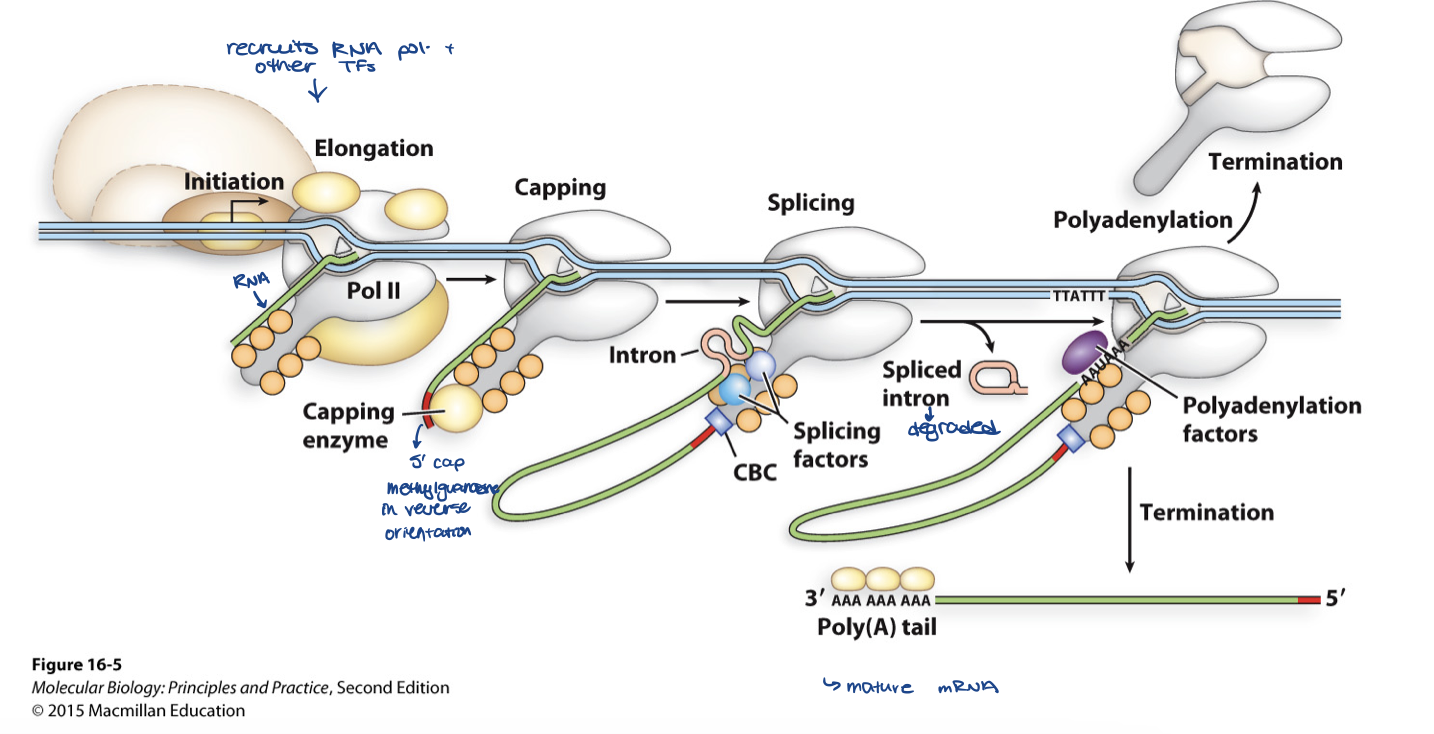
mRNA is modified in the ______
how is it modified?
nucleus
addition of cap to the 5ʹ end
splicing to remove the introns
requires breakage of the exon–intron junctions and joining of the ends of the exons
addition of A’s to 3ʹ end
RNA editing
what determines which introns are being spliced out?
expression of diff. small nuclear ribonucleoproteins (snRNPs) in diff cells and at diff times regulate which introns are being spliced
what happens to mature mRNA
it gets transported through nuclear pores cytoplasm where it gets translated
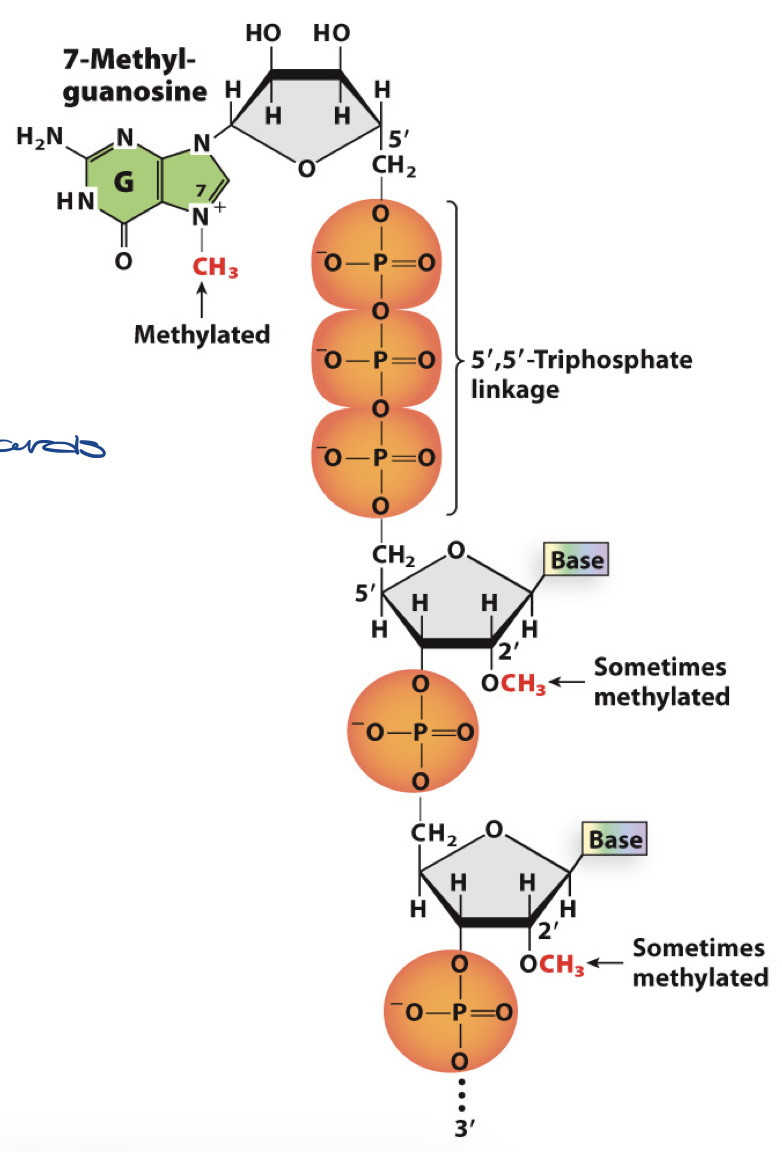
explain the addition of cap
add 7-methylguanosine (7-meG) - the 5’ cap
G is methylated
attachment is 5’ to 5’ phosphodiester bond
means added backwards
functions of 5’ cap
adds to stability of RNA
used as identity marker for ribosome to recognize
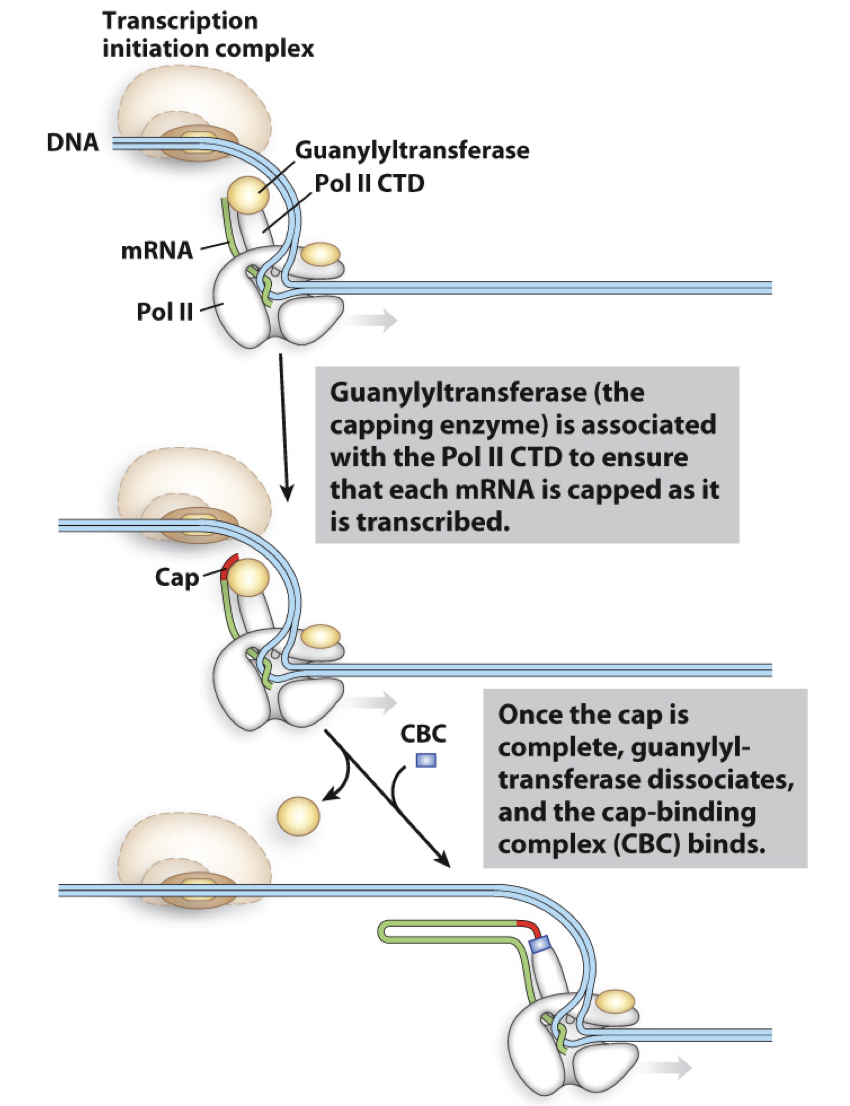
capping enzyme
enzyme - guanyltransferase
attached to CTD domain (phosphorylated) of pol II to be sure every RNA transcribed by RNA pol II is capped
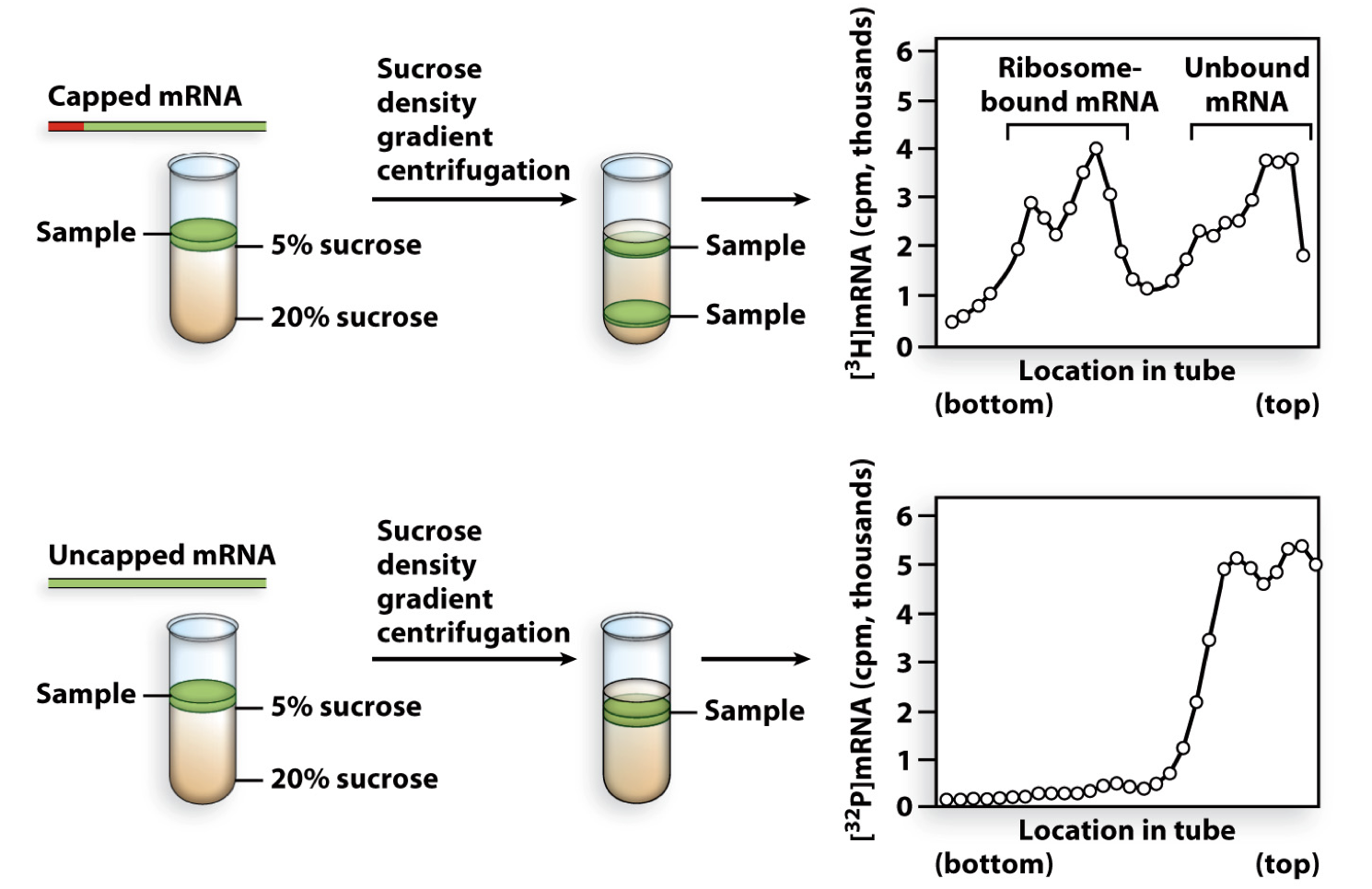
how do we know 5’ cap is necessary?
experiment showed that capped mRNA gets bound to ribosome
uncapped mRNA does not bind ribosomes
therefore the 5’cap is necessary for ribosome binding
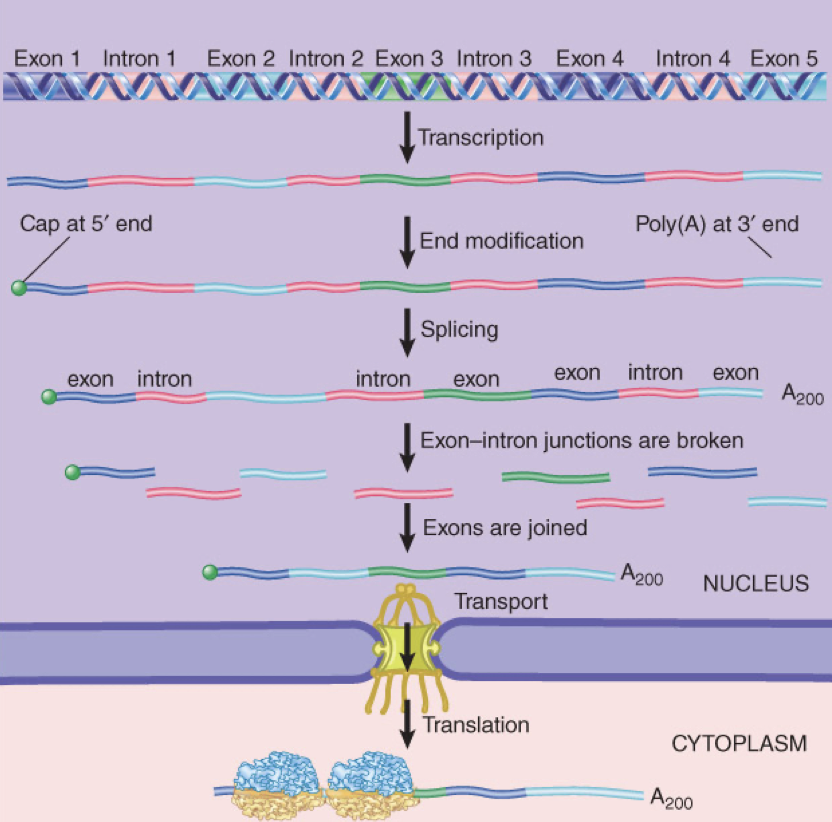
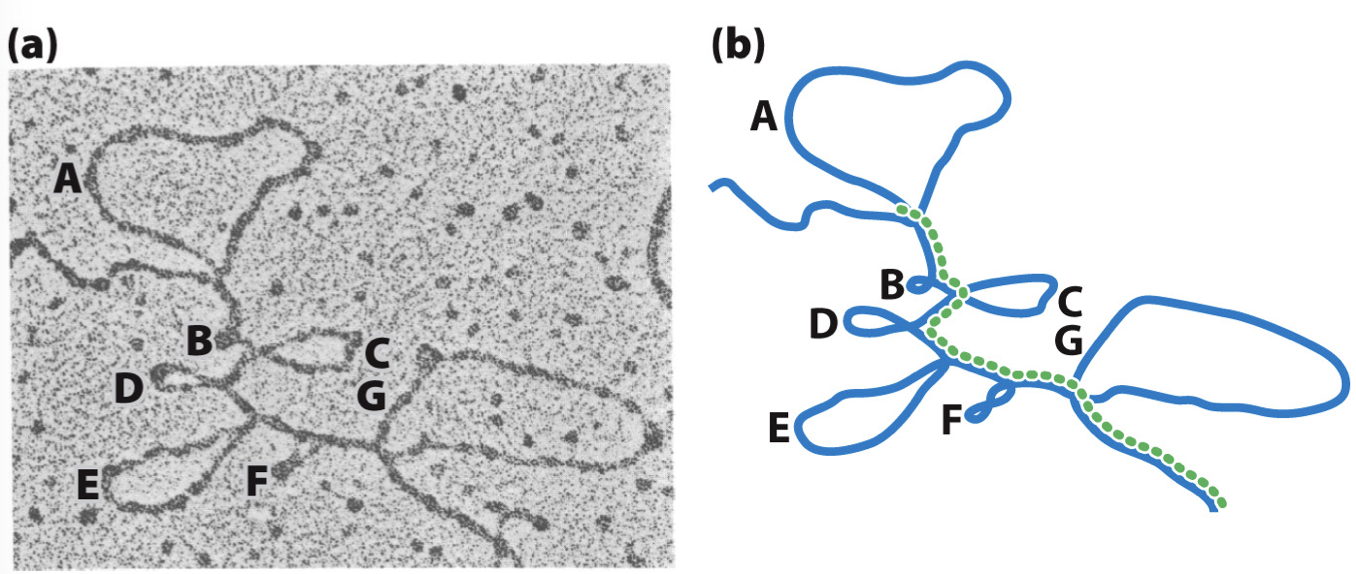
how do we know there are introns in DNA?
hybridized mRNA and corresponding DNA sequence has loops
e- microscopy shows loops which indicate where the introns are
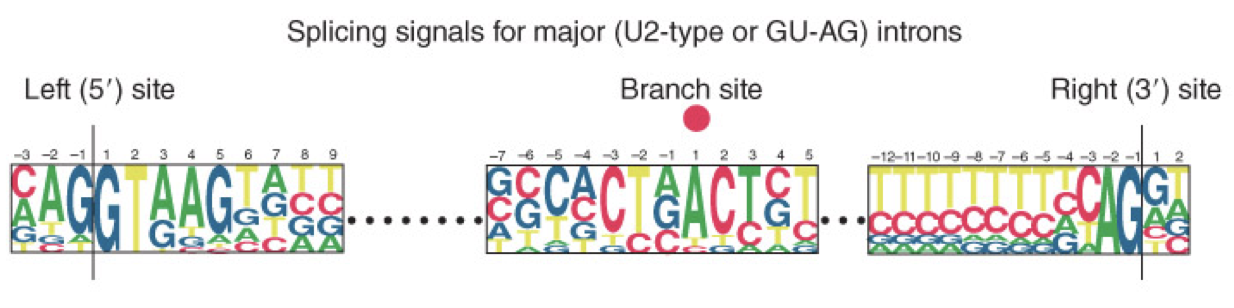
GU-AG Rule (U2 Type)
seen in majority of introns
defined the requirement for these constant dinucleotides at the first 2 and last 2 positions in the intron
the ends of nuclear introns are defined by the GU-AU rule (seen as GT-AG in the DNA sequence)
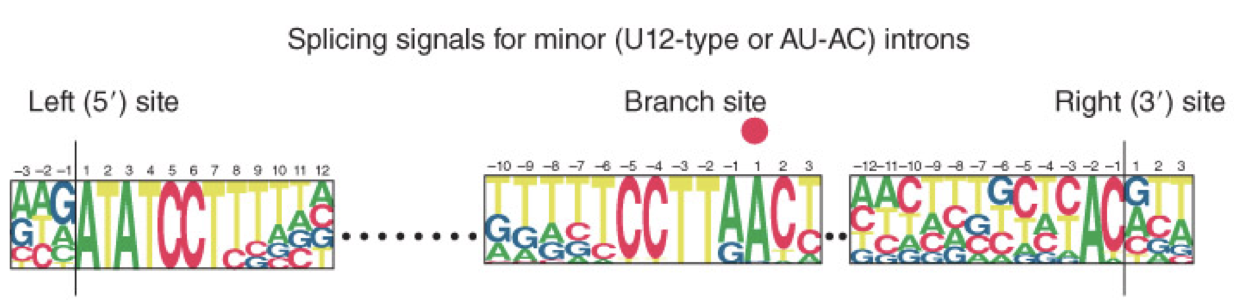
AU-AC Rule (U12 Type)
observed in a minority of introns
minor introns are defined by different consensus sequences at the 5’ splice site, branch site and 3’ splice site
what do these rules ensure
accurate and regulated splicing of pre-mRNA, allowing for generation of mature mRNA molecules
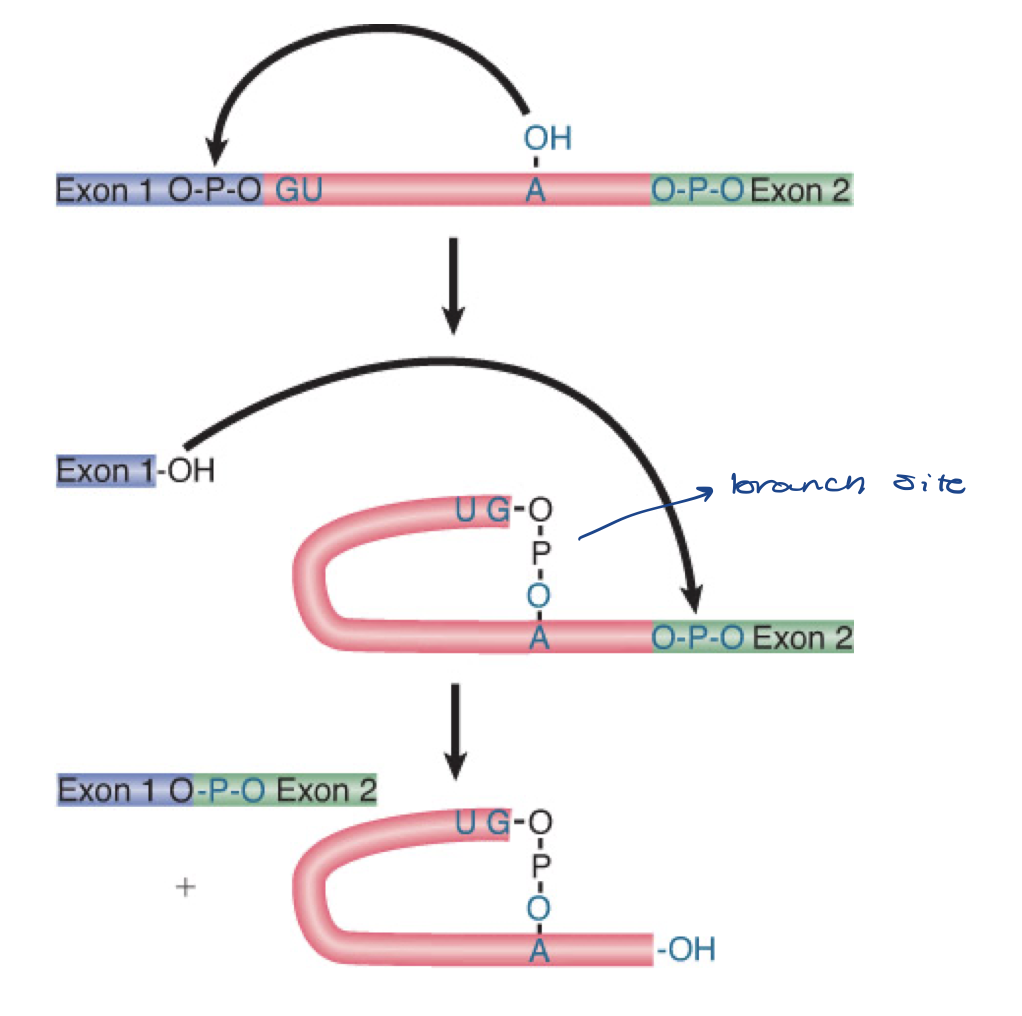
splicing occurs in two stages, describe them
5ʹ exon is cleaved off, then it is joined to the 3ʹ exon
adenine (A) serves as branch point
nuclear splicing occurs by two transesterification reactions, in which an -OH group attacks a phosphodiester bond
carried out by spliceosome
intron gets released
what do spliceosomes recognized?
5’ splice site
branch site
3’ splice site
spliceosome composition
approximately 12 megadaltons (MDa)
complex of RNA and proteins
five snRNPs account for almost half of mass
remaining proteins include known splicing factors and proteins involved in other stages of gene expression
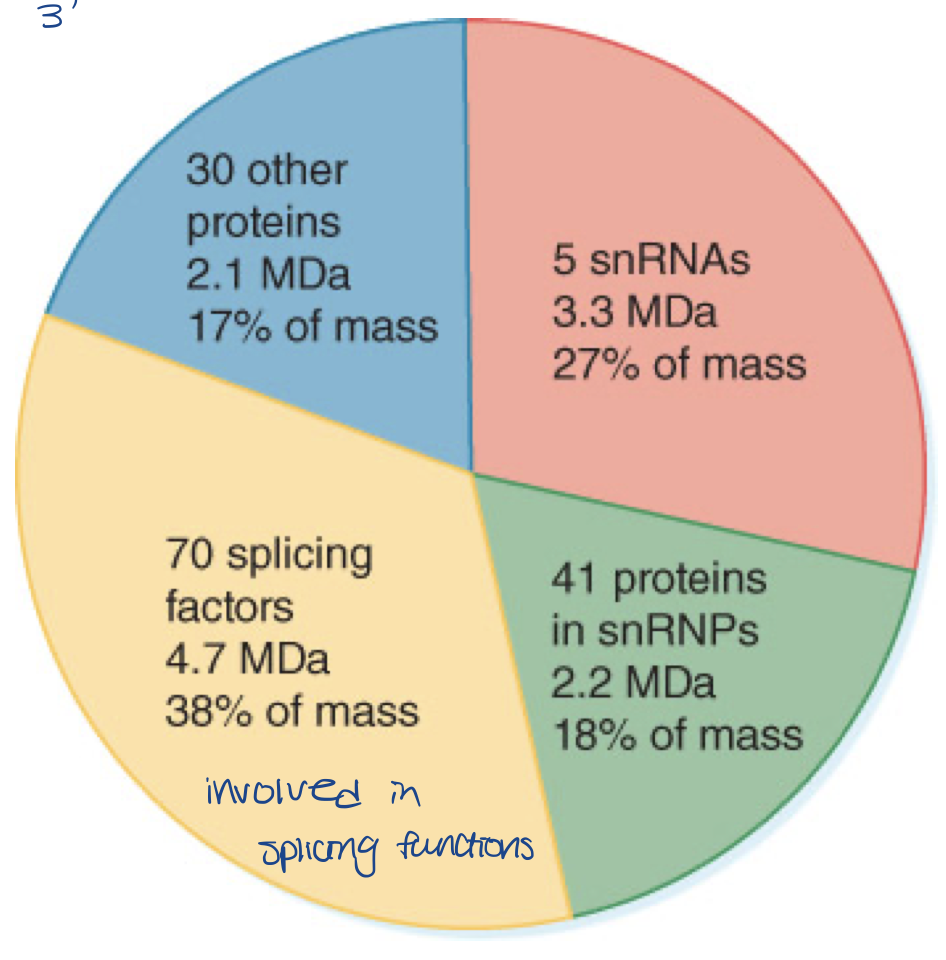
how many snRNPs in spliceosomes
5 snRNPs (snurps)
U1, U2, U4, U5 and U6 (not for memory)
plus hundreds of additional proteins
AND one small nuclear RNA (snRNA) at the heart of each snurp
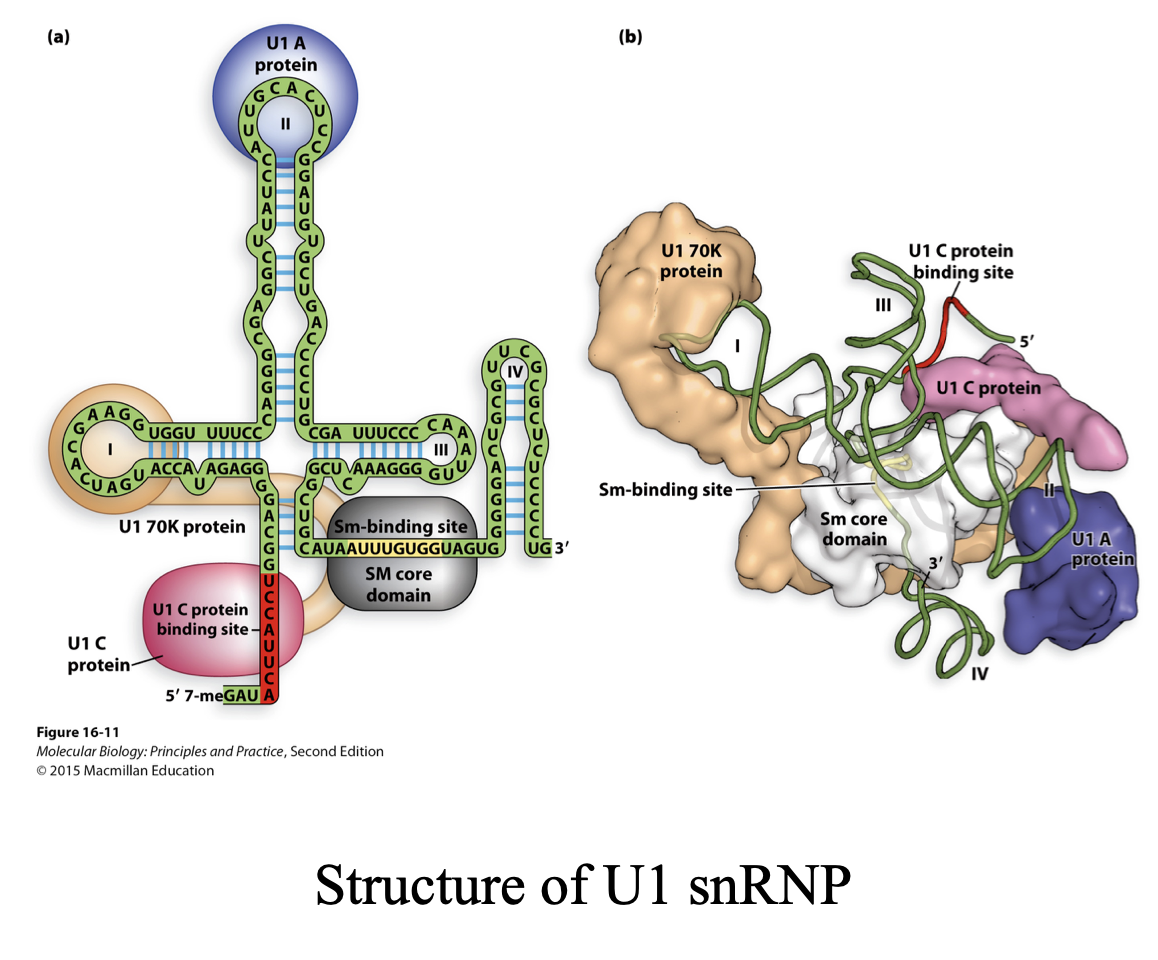
U1 small nuclear RNA (snRNA)
describe the structure
base-paired structure that creates several domains.
5ʹ end remains single stranded and can base pair w/ 5’ splice site
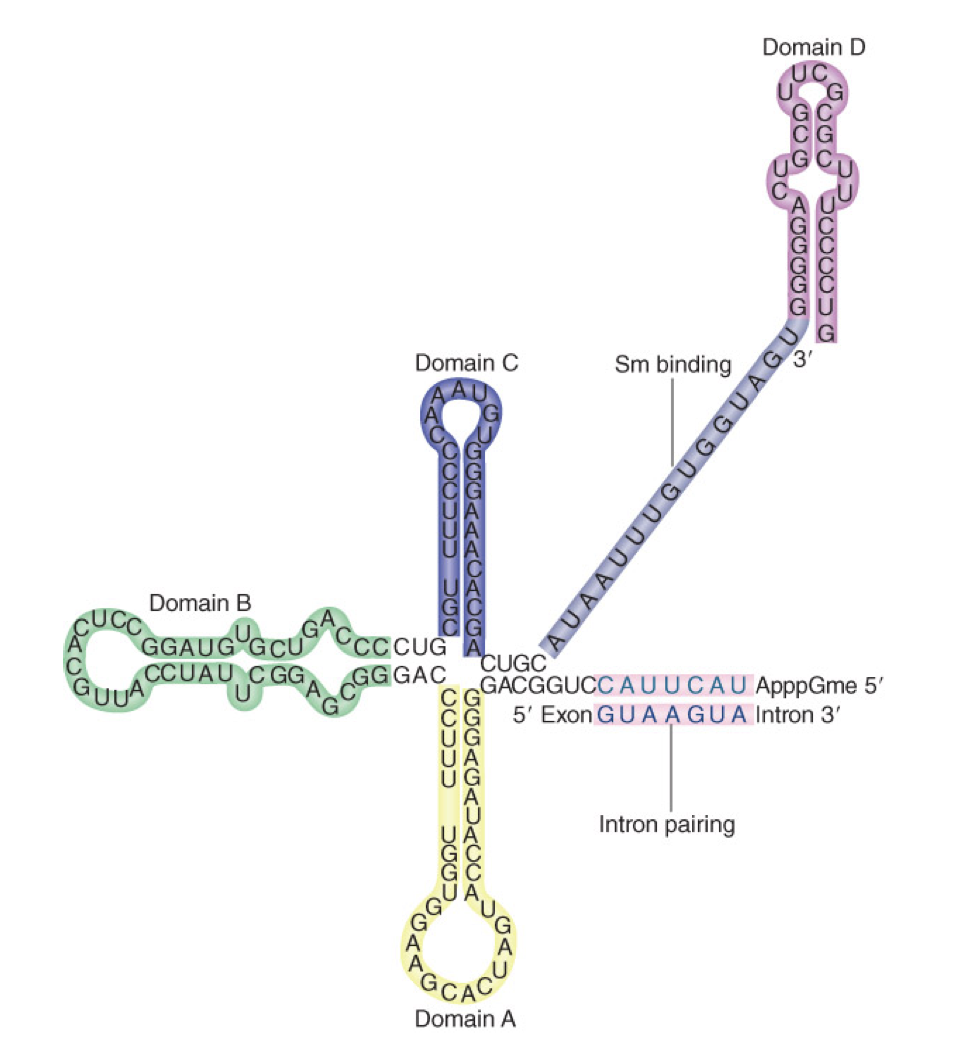
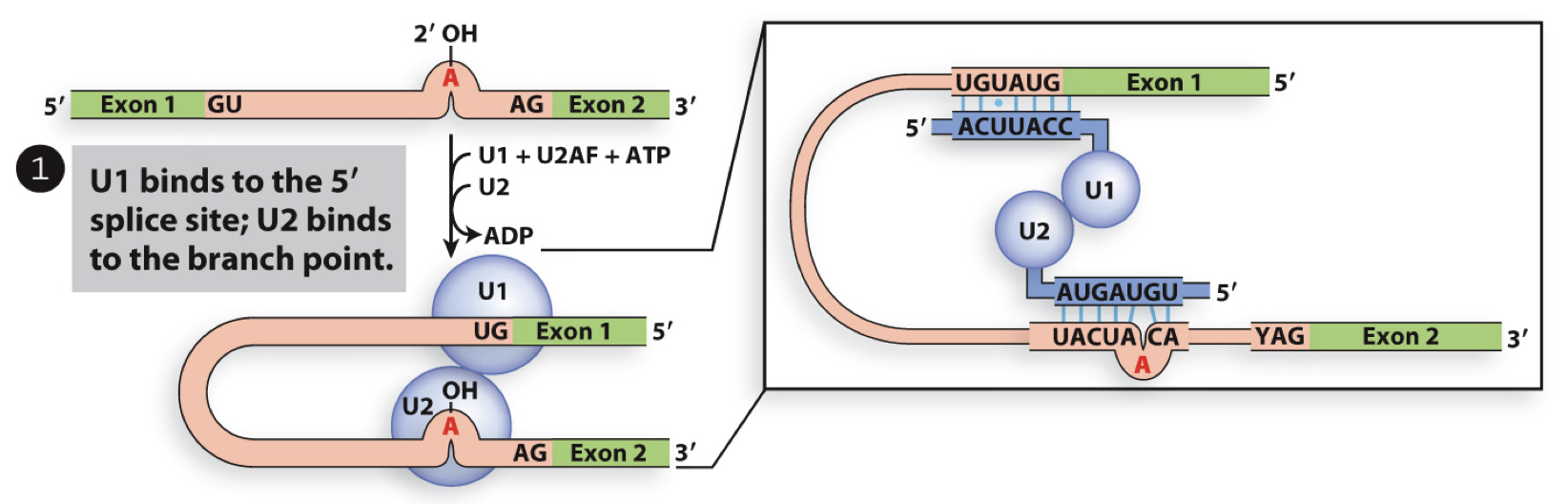
U1 in splicing - step 1
U1 binds to GU at 5’ splice site and U2AF binds to AG at 3’ site and helps U2 bind to branch point (A)
requires energy (ATP hydrolysis)
U1 in splicing - step 2
2’OH at branch point then becomes a nucleophile that attacks the phosphodiester bond at the 5’ splice site
U4-U6-U5 trimeric snRNP complex displaces U1 at the 5’s splice site then U4 dissociates
U1 and U4 snRPS are released
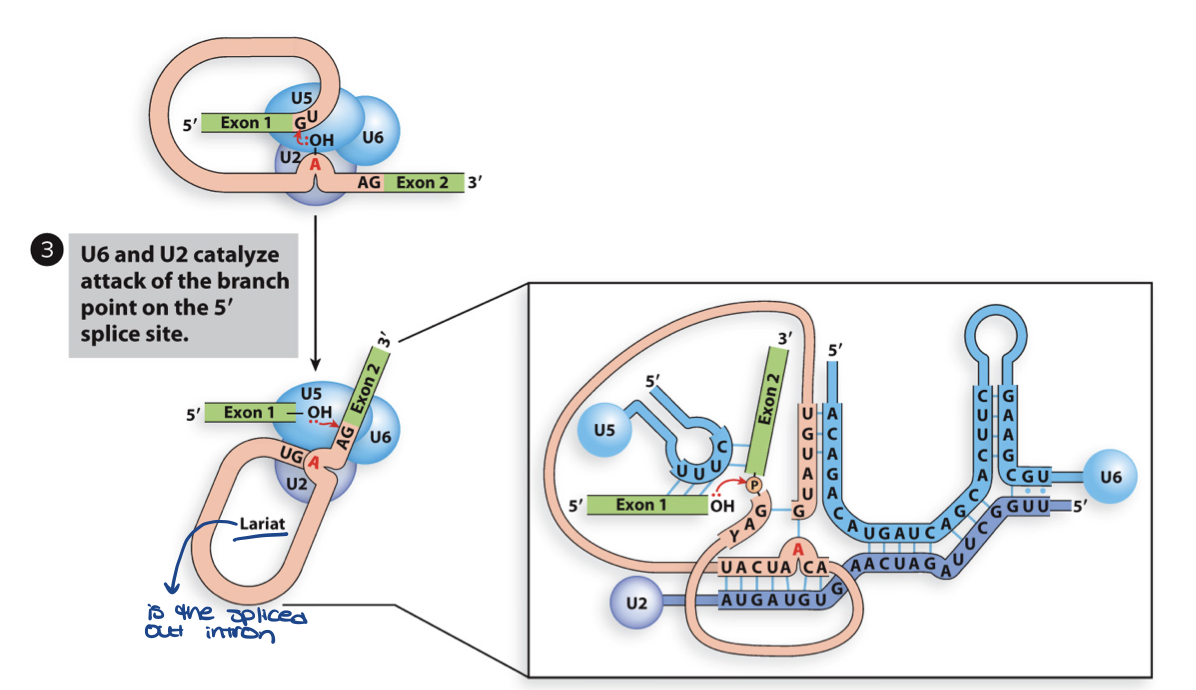
U1 in splicing - step 3
U6 and U2 catalyze n-philic attack of 2’OH branch point A on phosphodiester bond at the 5’ end – cleaving the 5’ exon- intron junction
this causes U5 to shift to 3’ splice point
first step in transesterification
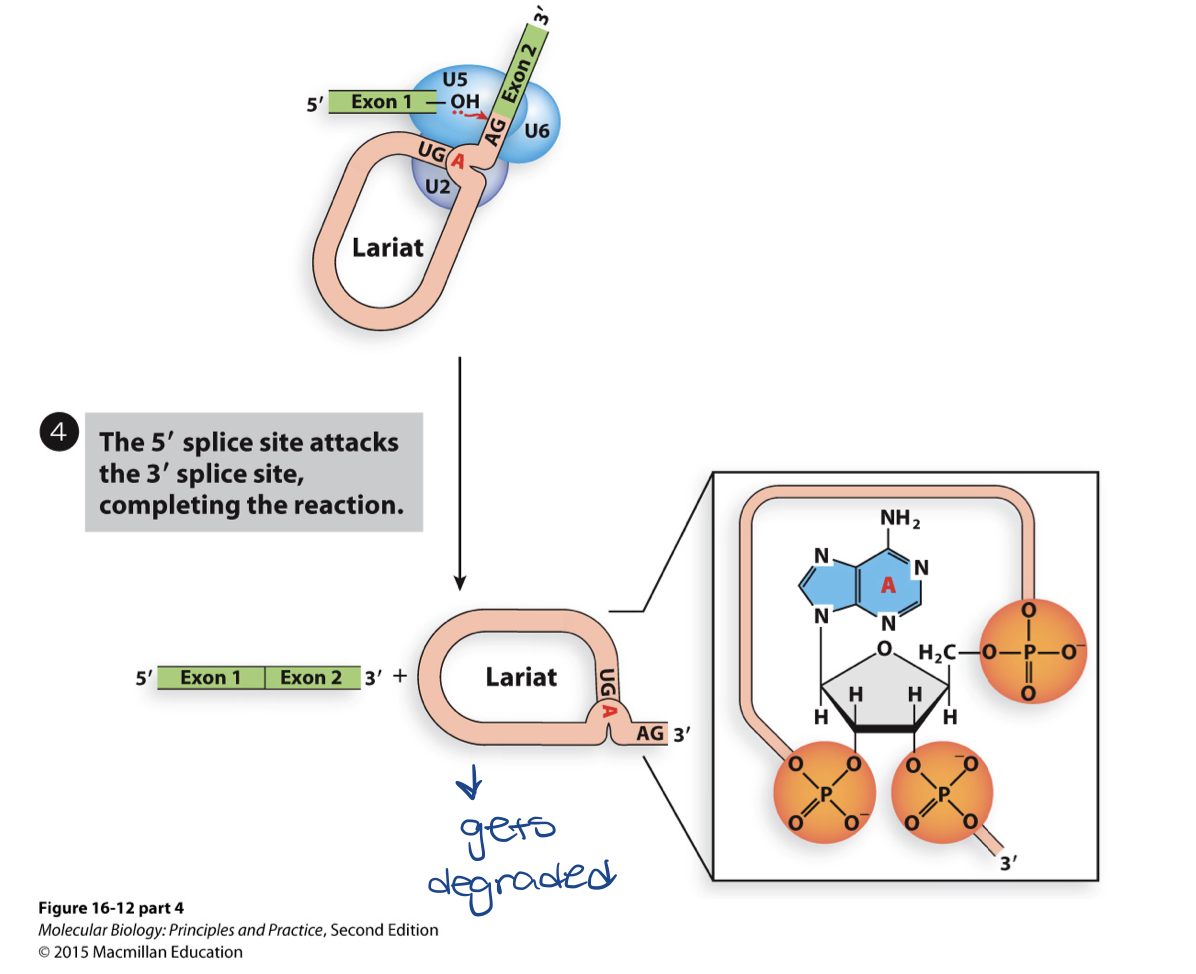
U1 in splicing - step 4
results in lariat (intron that’s cut out in a lasso/non-linear shape) containing the intron attached by the 3’ end
U2-U6-U5 remains attached and causes nucleophile attack on phosphodiester bond at 3’end
results in release of intron and joining of 5’ and 3’ ends on exons
requires energy
second step in transesterification
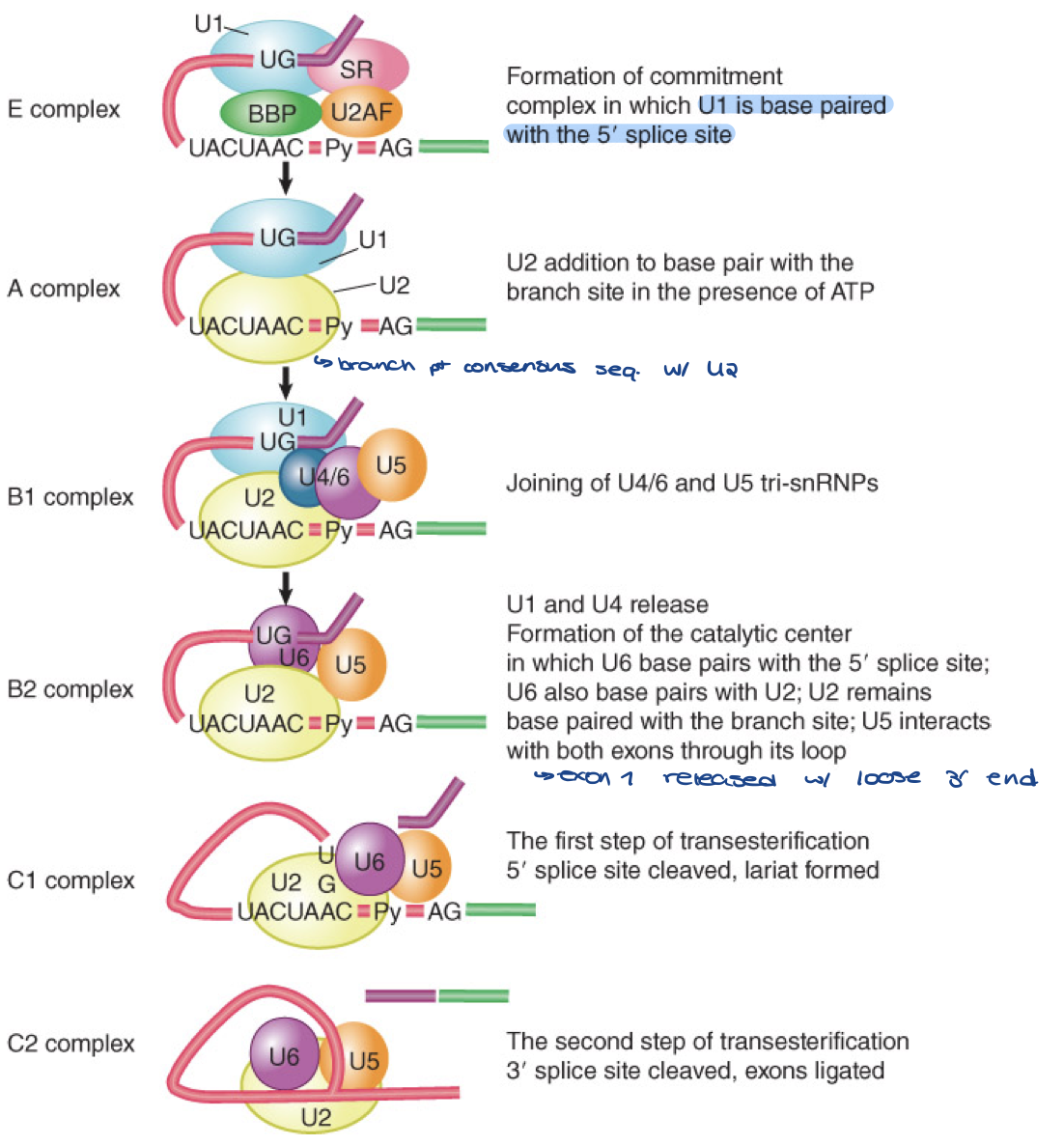
summary
splicing reaction proceeds through discrete stages in which spliceosome formation involves the interaction of components that recognize the consensus sequences
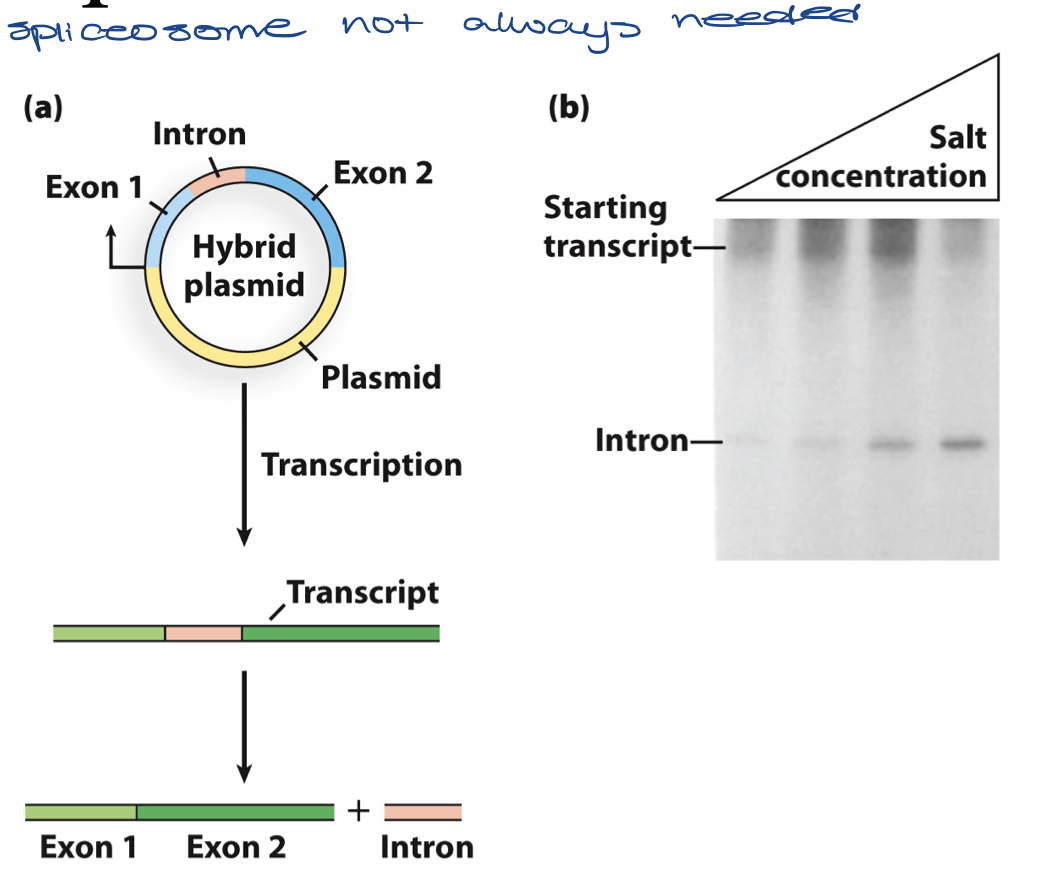
describe the experiment that showed that some introns can self-splice (no need for spliceosome)
eukaryotic gene cloned into bacterial plasmid vector and transcribed with bacterial RNA polymerase in vitro
found it had spliced itself (without help from any proteins)
reveals that RNA is capable of enzymatic activity
name and describe the 2 classes of self splicing introns
group I introns
always self-splice
share a common structure
group II introns
usually use a spliceosome (as a catalyst) but are capable of self-splicing
share a common secondary structure
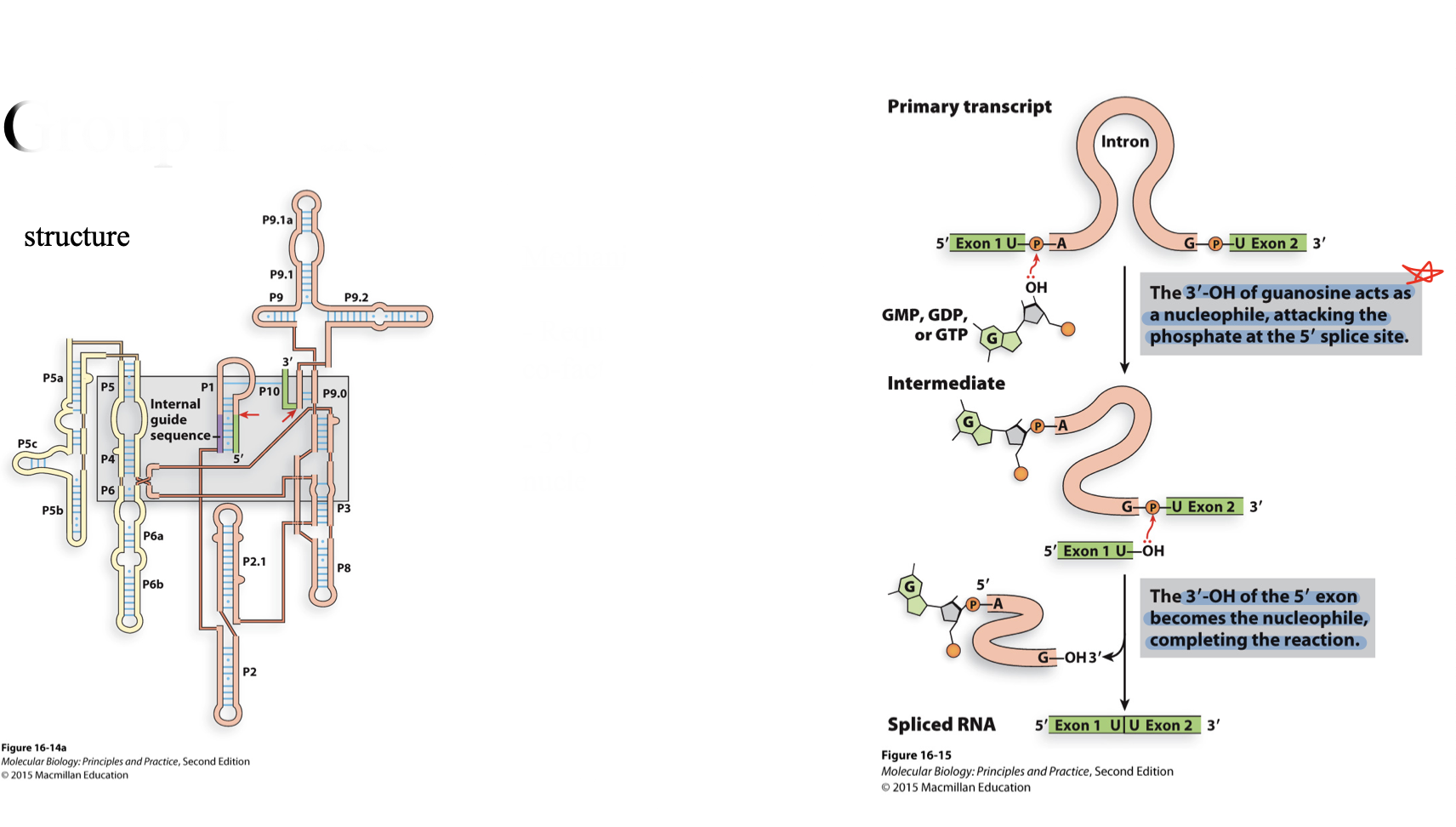
group I introns
structure
mechanism
lot’s of stem loops to bring exons together
requires a GTP as a co-factor not for energy
3’ OH used as nucleophile
3’ OH of guanosine acts as a nucleophile and attacks the phosphate at the 5’ splice site
3’ OH of the 5’ exon becomes the nucleophile and completes the reaction
exons are joined
intron is spliced out in a linear form
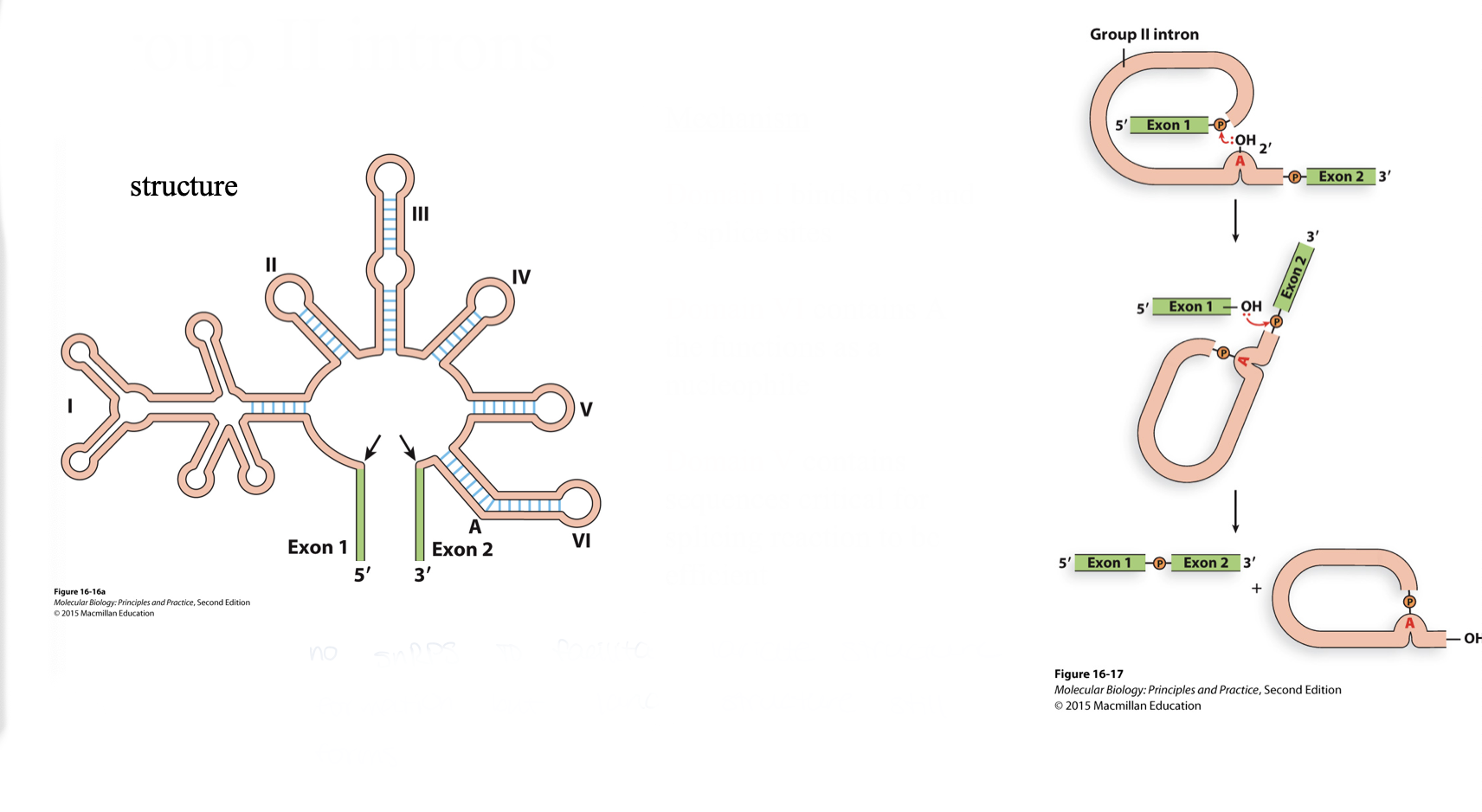
group II introns
structure
mechanism
lot’s of stem loops to bring exons together
domain I binds to 5’ and 3’ splice sites
domain VI contains A the functions as a nucleophile
domain V contains sequences critical for splicing reaction to be efficient
lariat structure forms
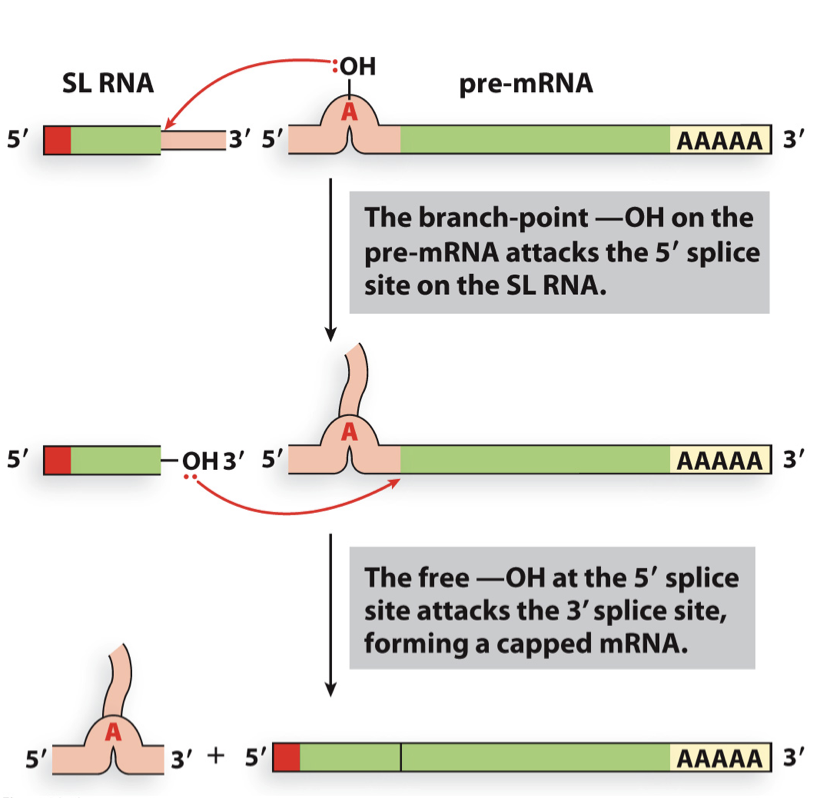
trans-splicing
occurs in nematodes (conserved in that group)
short capped leader sequence is spliced onto a pre-mRNA to make a mature mRNA
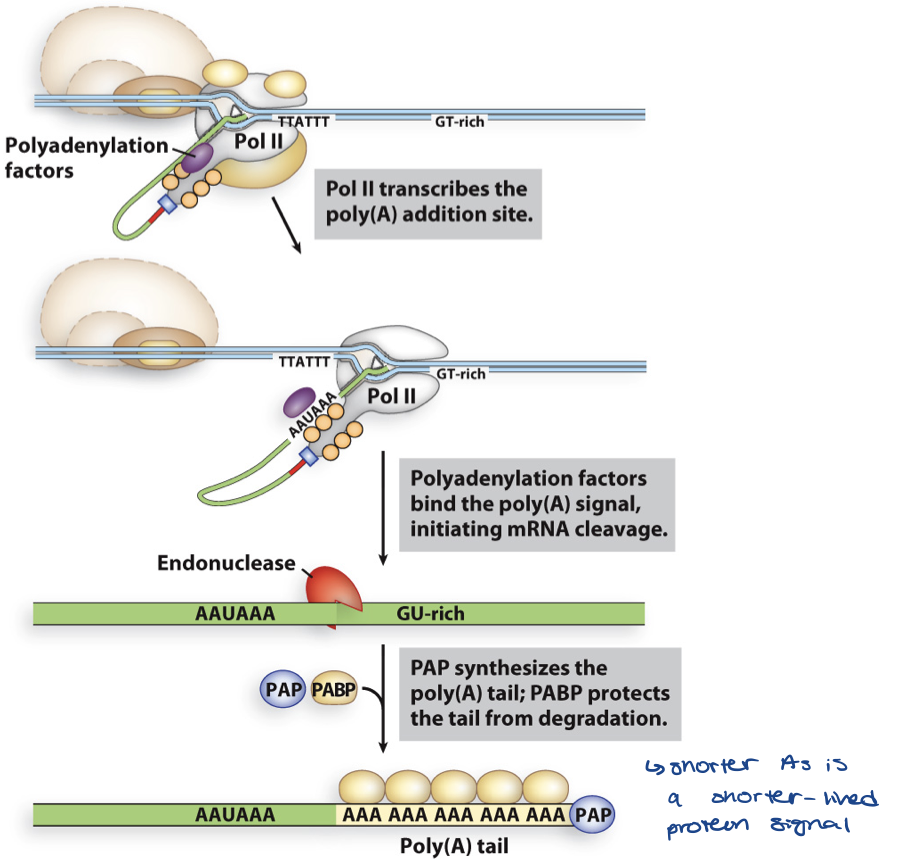
adding poly A tail
length
function
typically 80–250 residues long
length determines how long mRNA lasts
function:
stability by blocking access of ribonucleases to 3’ end
mRNA capping, polyadenylation and intron splicing
all three are coordinated