Molecular biology techniques - molecular cloning, PCR, g
1/96
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
97 Terms
What is the point of molecular cloning?
assemble recombinant DNA
Define recombinant DNA
DNA coming from more than one source, has been spliced and recombined with synthetic dna
What’s the point of cloning DNA? (Give 2)
isolate a region of DNA
sequence a gene
Analyse mutant vs WT genes
Make mutations in the DNA
Express and purify the protein
What 2 physical elements are needed to create recombinant DNA?
a vector and an insert
What are the 3 steps of molecular cloning?
creation of recombinant DNA
transformation: transfer of dna to host
selection of the hosts carrying our inserts and replicating them
What is the role of a vector?
carrier of selected gene
What makes a vector better suited than a DNA insert alone?
Insert DNA can probably get into a host cell – but it won’t last long there
Vector has features that enable it to get into a host and maintain / replicate itself there
Anything DNA that is part of the vector will be maintained / replicated too
What is the most commonly used vector?
a plasmid
What are the 3 components of a vector?
selectable marker
Multiple Cloning Site
Origin of replication
What’s the function of the selectable marker of a vector?
allows survival of host cells that are carrying your plasmid only
What’s the function of the multiple cloning site of a vector?
Where the gene is cloned onto the vector + has restriction enzyme sites
What’s the function of the origin of replication of a vector?
Independent replication inside the host allows the insert to survive over multiple generations
By what is DNA cut in a specific place?
Restriction enzyme/Restriction endonuclease
What is a restricted phage?
phage grown in a first host failed to be passed on through other generations
What is a modified phage? How is it protected from degradation?
phage grown in a first host managed to be passed on through other generations
phage acquires the correct methylation pattern
How many types of restriction enzymes exist? Which ones are used to cut DNA at a defined position?
4
type II
Where do type II restriction enzymes cut DNA in relation to the recognition site of DNA?
either within or near it
How are restriction enzymes named?
First 3 letters = species (E. coli)
4th letter = strain (RY13)
Roman numeral = which enzyme from that species/strain (at least 5!)
Are the most used type II restriction enzymes heterodimers or homodimers? Define the term.
homodimers
two identical polypeptides
Is DNA palindromic? Define the term and explain how that impacts reading of both branches.
yes = same backwards and forwards
reads same 5’-3’ on each strand.
Can type II REs recognise specific sequences?
yes
How many base pairs on average do REs recognise?
4-8 (6 is most common)
Do type II REs generate sticky/overhang ends or blunt ends? Define both terms
Both:
overhang = an uneven cut where there are a few base pairs that are unpaired at the end of the molecule (5’ or 3’ end)
blunt end = clean cut where all bp are paired
What does the final cleavage of a fragment of DNA create at the 5’ and 3’ ends respectively?
5’: phosphate group
3’: OH group
Name a specific Restriction Enzyme that results in blunt ends after cutting?
SmaI
Name a specific Restriction Enzyme that results in overhang/sticky ends after cutting?
HindIII
What are the 5 steps that occur that result in the cutting of DNA?
Initial binding is non-specific.
Enzymes moves along DNA until it finds a specific recognition site.
Specific binding triggers structural changes (enzyme and DNA)
Catalysis requires Mg2+.
Generates free 5’-phosphate and 3’-OH ends.
Which ion is a co-factor in the process of cutting DNA?
Mg2+
When cutting overhanging ends in DNA sequence of the vector and the insert, should one single type of enzyme be used or several in order to ensure compatibility between the overhanging ends? Why?
ONE SAME TYPE. If two different enzymes cut the vector and the insert respectively, they are likely not going to result in the formation of compatible overhanging ends.
Does the compatibility of overhanging ends suffice to hold the insert and vector together? Why? (Which bonds are created between base pairs)
no, creation of hydrogen bonds (and not covalent) between pairs is too weak to hold then together for long
Which enzyme allows a permanent linkage of vector and insert? How ?
DNA ligase
formation of phosphodiester bond in the sugar phosphate backbone of the dna sequences, linking them together
What structure of overhanging ends allows ligase to create a phosphodiester bond in the sugar phosphate backbone of the vector and insert dna sequences?
phosphate and OH groups
Is hydrogen bonding between complementary bases more efficient at lower or higher temperatures?
lower
Does ligase form the phosphodiester bond before or after the formation of a hydrogen bond between bases?
After! this bond allows ligase to bind the ends more efficiently
Is ligase more efficient at lower or higher temperatures?
higer, around 20 degrees C
Can ligation occur between blunt ends ? Is it more or less efficient than with overhanging ends? Why?
yes
less efficient because there’s no temporary hydrogen-bonding between complementary ends so they’re not already temporarily bound
Define a restriction site
specific sequence of DNA nucleotides that restriction enzymes can identify and cut
What potential issues could arise in molecular cloning? (2 ex.)
no convenient restriction sites
not have enough DNA
DNA mixed in with lots of other DNA molecules
vector self ligating
gene inserted in wrong orientation
Why is vector self ligation probable ? How can it be avoided?
bc ends are complimentary and close together even after cutting
phosphatase treatment removes 5’ phosphate – no phosphodiester bond can be formed
Is molecular cloning 100% effective?
NO
What happens if only one of the two DNA strands has a phosphate group? How is it resolved in the host cell?
no phosphate = no ligation so the lacking strand would not form a phosphodiester bond = nick in DNA
repaired by host enzymes once inside a bacterial cell
Name an example of a type of DNA that would require phosphorylation.
PCR product
Why does DNA need to be phosphorylated when lacking a phosphate group on its 5’ end ? Which enzyme would do this and does it require energy?
for ligation to work
T4 Polynucleotide kinase, yes ATP
What is done more often: addition or removal of a phosphate group to 5’ ends? Why?
removal to avoid self ligation of the vector
Which enzyme removes the phosphate group from the 5’ end of vector’s DNA sequence? Can ligation still occur?
Calf Intestinal Phosphatase (CIP)
no, just hydrogen bond pairing
What’s the intention behind removing a DNA overhang?
Blunt end cloning might be necessary
Destroy restriction enzyme sites
What are the 3 ways of removing a DNA overhang?
fill in the 5’ overhang
remove the 3’ overhang
remove the 5’ overhang
What is mung bean nuclease responsible for?
removing the 5’ overhang
What are T4 DNA Polymerase or DNA Polymerase I responsible for?
filling in the 5’ overhang and removing the 3’ overhang
Can the 3’ overhand be filled?
no
What is the most common way of transferring the recombinant DNA to the host cell?
using a bacteria called E. coli
What are 2 ways of getting the recombinant DNA through the membrane of the host cell?
Electroporation uses high-voltage – thought to induce transient pores in cell membranes
Chemical transformation of E coli + heat-shock – membrane changes
What is an example of a selectable marker? How does it work?
specific antibiotic resistant gene
only hosts with the new recombinant DNA will have the selectable marker gene that is antibiotic resistant allowing it to survive in an antibiotic rich env., the others will die
As modified hosts divide, does the vector also divide? Thanks to what part of the vector?
Yes hence the term molecular cloning
thanks to the origin of replication part
What does PCR stand for?
Polymerase Chain Reaction
Is DNA negatively or positively charged? How does this explain how electrophoresis works?
negatively
charge is positive at the opposite end of the dips in gel = migrating DNA from minus to plus charges
Does heavier or lighter DNA migrate farther in gel electrophoresis?
lighter
What is the basic goal of PCR?
DNA replication
What are the 6 elements needed for PCR ?
Template – DNA that we want to amplify
DNA polymerase – copies DNA
Primers – DNA polymerases need a free 3’-OH to start. Provided by a primer. (Therefore, we need to know a bit about our sequence)
Deoxyribonucleoside triphosphates (dNTPs) – DNA bases to make the new DNA strand
Buffer – Correct pH and ions (MgCl2)
Thermocycler – maintains appropriate temperature for each stage of a cycle
How do we find the correct number of molecules obtained after a specific number of rounds of PCR?
2nb of PCRs
Is amplification of DNA during PCR and why:
linear
exponential
exponential bc doubles each time, if linear we would just add 2 at each cycle
Define a probe
a single-stranded DNA or RNA sequence that is used to identify specific DNA or RNA sequences in a sample
What are the 3 steps of PCR and their respective temperatures?
denaturation 95
primer annealing 55-65
primer extension 68-72
What does the denaturation step of PCR consist of?
very hot to separate the two strands of DNA into 2 single stranded DNAs
What does the annealing step of PCR consist of?
Primers bind to complementary sequence on ssDNA. Primer binding is antiparallel so 5’ binds to 3’ and 3’ to 5’
What does the extension step of PCR consist of?
DNA polymerase synthesises new strands of DNA from the 3’ end of the primers
What is the main advantage of Taq and a disadvantage? What type of protein is it?
thermostability very high
no proof reading + A overhang
DNA polymerase
What is the main advantage of Pfu and a disadvantage? What type of protein is it?
thermostability very high + blunt ends
slower extension rate and processivity (efficiency)
DNA polymerase
What are 4 features of PCR primers ?
size : 17 is min, 20 is average
pairs: one on each strand of DNA in opposite directions, 3’ ends point towards each other
melting temp.
complementary specificity to template strand
binds in antiparallel fashion
What’s the difference between the forward and reverse primers? A similarity?
forward: binds to bottom strand of DNA in a 5’ to 3’ direction
reverse: binds to top strand of DNA in a 3’ to 5’ direction
bind in antiparallel fashion
Why and how is annealing temperature based on the primer’s melting temperature?
Bc don’t want to melt primer, annealing temp. should be about 5 C lower than primer melting temp. (so primers usually have about 60-64°C. Tm)
What happens to primers when the annealing temperature is too low?
primers may bind non-specifically to other DNA sequences
What happens to primers when the annealing temperature is too high?
primers may not bind efficiently (or at all), reducing product yield
What happens to primers when the annealing temperature is right?
primers will bind to your specific sequence
How is the melting temperature identified for G/C and A/T? What could explain the difference?
G/C: add 4°C
A/T: add 2°C
Different strength of bonds - 3 for GC, only 2 for AT
Roughly how many recombinant DNAs would we have after about an hour of PCR?
order of billions
What’s the issue with ligating a PCR product directly into a vector? What are 2 solutions that don’t depend on primers?
it has no 5’ phosphate so can’t directly bind to the 3’ OH of the vector
make a primer with phosphate or add a 5’ phosphate with T4
How effective is blunt ended cloning?
No, not directional
What is termed the business end of a primer? How does this affect the incorporation of restriction sites into primers?
the 3’ end, nothing can be added to it
when adding restriction sites to primers, this has to be on the 5’ end
Are restriction sites inserted into primers specific and bound to the template gene?
No
A primer is separate from DNA, by the end of PCR, is it incorporated into the DNA sequence? How?
yes
copied during PCR
How is the issue of ligating a PCR product into a vector solved by primers?
ligation allowed by sticky ends of the product (the primers) + no self ligating of vector and made directional through different enzyme interactions
What is RT-PCR? Which organisms do it naturally? What does it refer to?
reverse transcriptase polymerase chain reaction
viruses
RNA made into cDNA
How is the first strand of DNA made during reverse transcription?
poly(dT) primer binds to the mRNA’s poly(A) tail
synthesis of first strand with reverse transcriptase and dNTPs
RNA is removed
How is the second strand of DNA made during reverse transcription?
DNA polymerase I (Klenow fragment) synthesises second strand from the hairpin structure left over from first strand - acts as a primer
the loop is then digested by a nuclease
When does PCR stop being exponential?
when reagents run out, after about 30 rounds of PCR or DNA polymerase lose activity
Is qPCR done during the exponential or plateau phase of PCR? What does it measure?
exponential
measures/quantifies the amount of DNA made from PCR
How is fluorescence used in qPCR? What’s a limitation of the technique? Give an example of a fluorescent protein marker
Fluoresces when it binds dsDNA but not sequence specific so could be any DNA
SYBR Green
How are fluorescent probes used in PCR?
Sequence specific, once the probe attached to DNA is cleaved, snaps in to and becomes fluorescent
What is cycle threshold? Is the measure relative or absolute?
point at which fluorescence exceeds background levels
relative - which one had most template at start
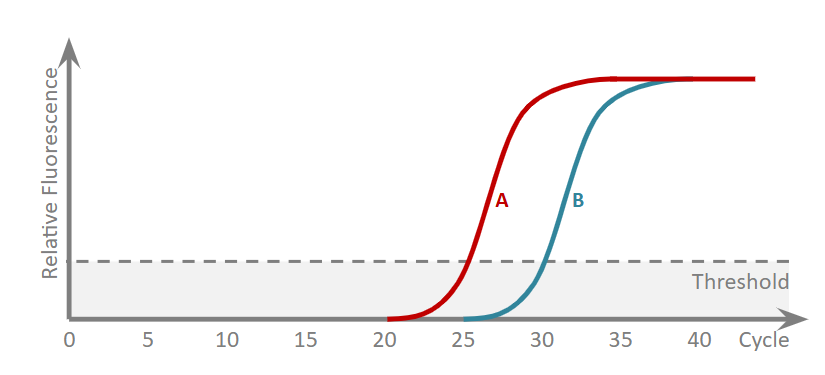
Does A or B have more template to start with? Why?
A, lower Ct = fewer cycles of PCR needed to exceed threshold = more template at start
Find the difference in Ct values, delta Ct
Ct A - Ct B
Find the fold difference
2−ΔCt - this gives how much more template A has over B
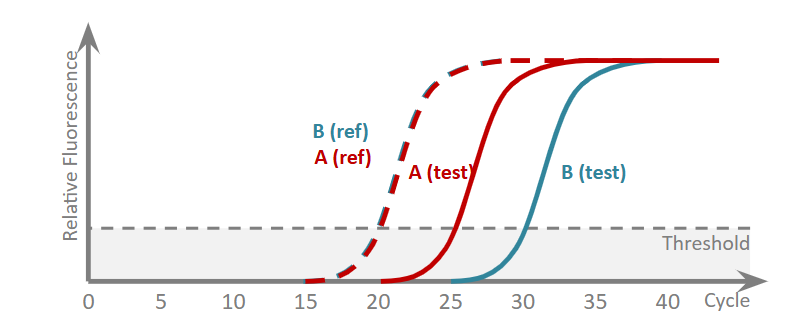
Why is this method the most accurate way to find the fold difference between A and B? How do you find it?
we have a more accurate value for the relative fluorescence
2−ΔΔCt where ΔΔCt = ΔCtA − ΔCtB
What are the 3 molecules that can be analysed to measure gene expression?
RNA
DNA
Proteins
How does PCR allow us to analyse the expression of our gene?
shows