Oxidative phosphorylation 19.1-19.3
1/34
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
35 Terms
Oxidative phosphorylation
e transfer reaction in mitochondrion
-outer mm-pores→ free diffusion of small molecules
inner mm -tight→ no free passage
e transfer between e carriers
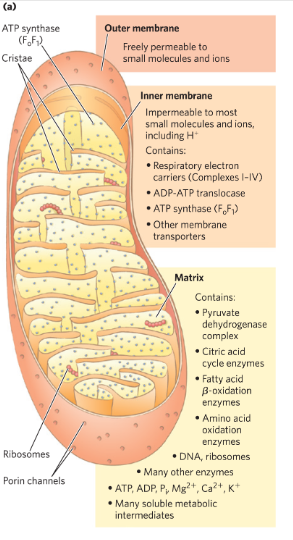
Mitochondrial matrix
Enclosed by an inner mm → contains pyruvate dehydrogenase complex + enzymes of the citric acid cycle, fatty ox pathway and amino acid ox → ONLY glycolysis which occur in the cytosol
Respiratory chain
Electrons enter a series of electron carriers - e are transported by NADH and FADH2
3 types of e transfer occur in ox phosp
Driect transfer of e as in Fe3+→ Fe2+
Transfer as a hydrogen atom (H+ +e-)
Transfer as a hydride ion (:H-) carrying 2 e-
Electron carrying molecules
NAD
FLavoproteins
Quionone - coenzyme Q
Cyrochromes
Fe-S proteins
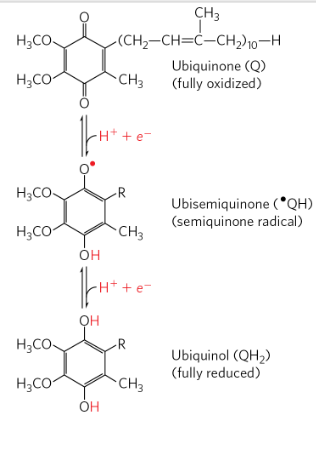
Ubiquinone Q
Lipid soluble benzoquionone with a long side chain
Can accept one e to become semiquinone radical
Or 2 electrons to becom ubiquinol
Freely diffusible in the lipid bilayer
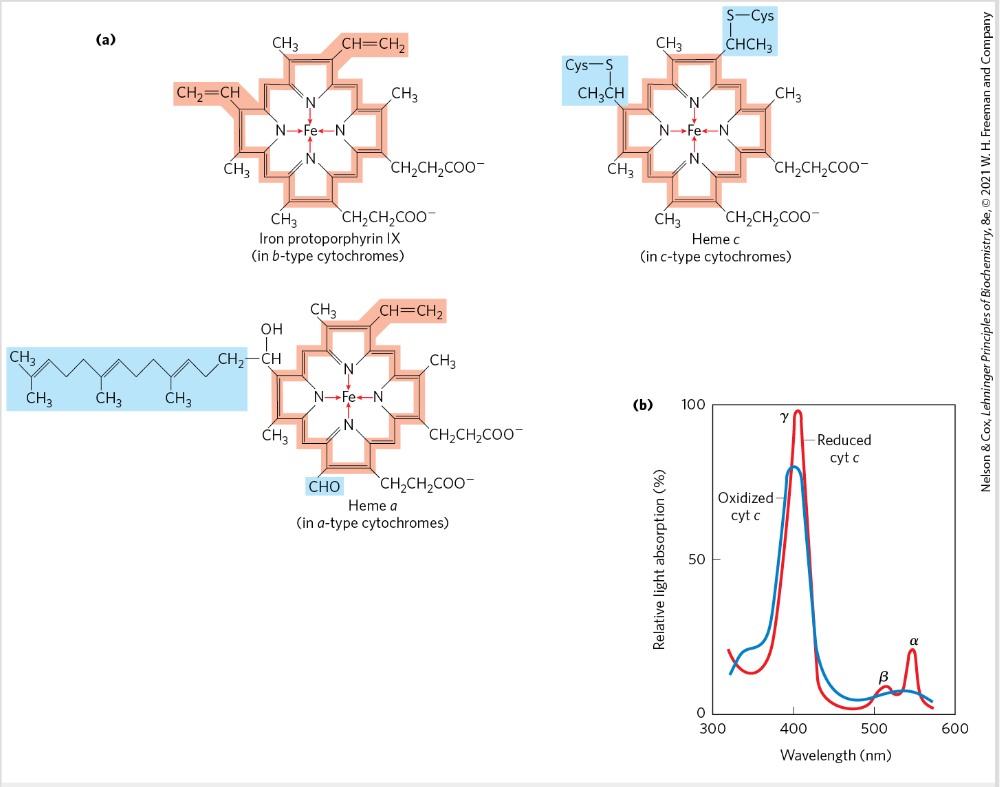
Cyrochromes
=proteins with characteristic strong absorption of visible light- heme groups
three classes: a, b and c → different light absorption spectra
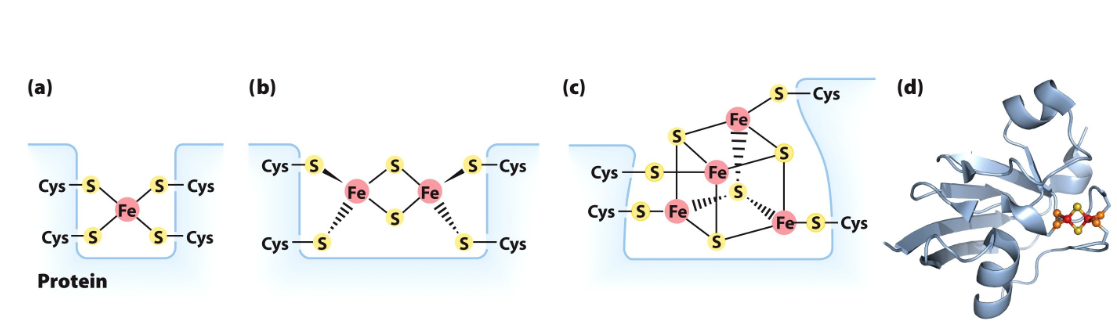
Fe-S proteins
Iron (not in heme) with sulfur atoms or with sulfur of Cys residues in the protein or both
Participate in one electron transfer- one iron atom of the cluster is oxidized or reduced
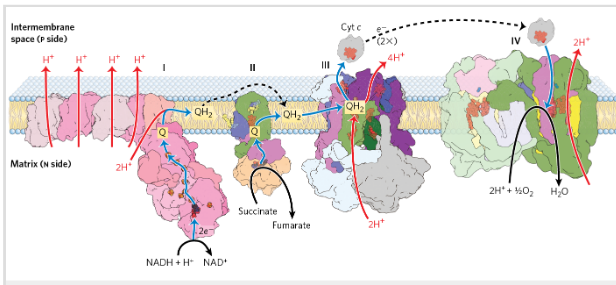
Resipratory chain electron carriers
E move from NADH, succinate, or other primary electron donor _> flavoproteins, W, Fe-S proteins and cytochromes to O2
Is e transfer favorable?
Yes! Its spontaneous:
NADH → Q → cytochrome b →
cytochrome c1 → cytochrome c →cytochrome a → cytochrome a3 → O2
E flows from carriers of low E’ to higher E’
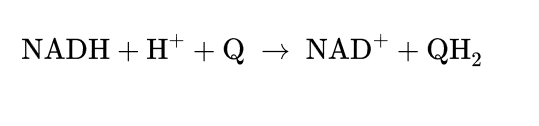
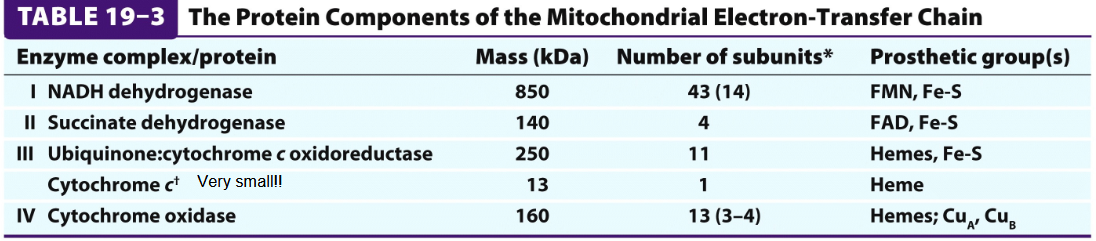
COmplex 1= NADH dehydrogenase
Catalyze transfer of hydride ion to Q from NADH (exergonic)
4 H+ is pumped to intermm space (endergonic) (against a proton gradient)
NADH+5H+ (N)+Q→NAD+ + QH2 + 2H+ (P)
Prosthetic groups= FMN containing flavoprotein, Fe-S
L shape
Proton pump is driven by e transfer
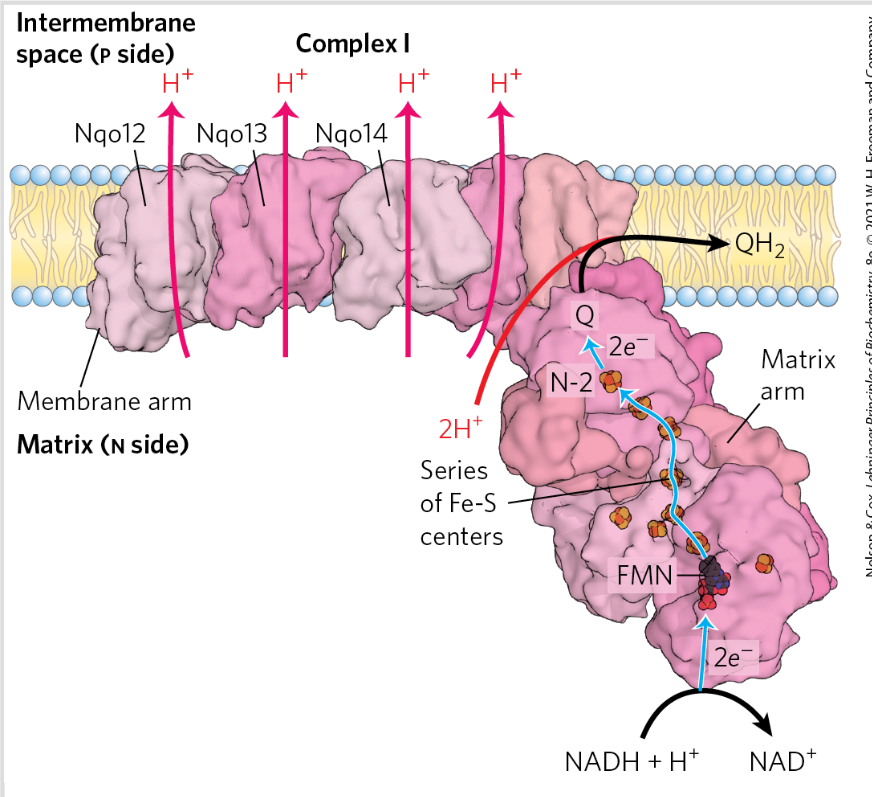
Complex 2=succinat dehydrogenase
Catalyze electron transfer from succinate (ox) to Q (reduced)
Prosthetic groups= FAD, Fe-S
NO proton pump
(Functions in the citric acid cycle)
2 transmembrane subunits C and D
COmplex 3= Ubiquinone: cyt c oxidoreductase/cyt bc1
Carries e from reduced Q to Cyt c
Prosthetic groups: Heme, Fe-S
Proton pump
Dimer- functional unit
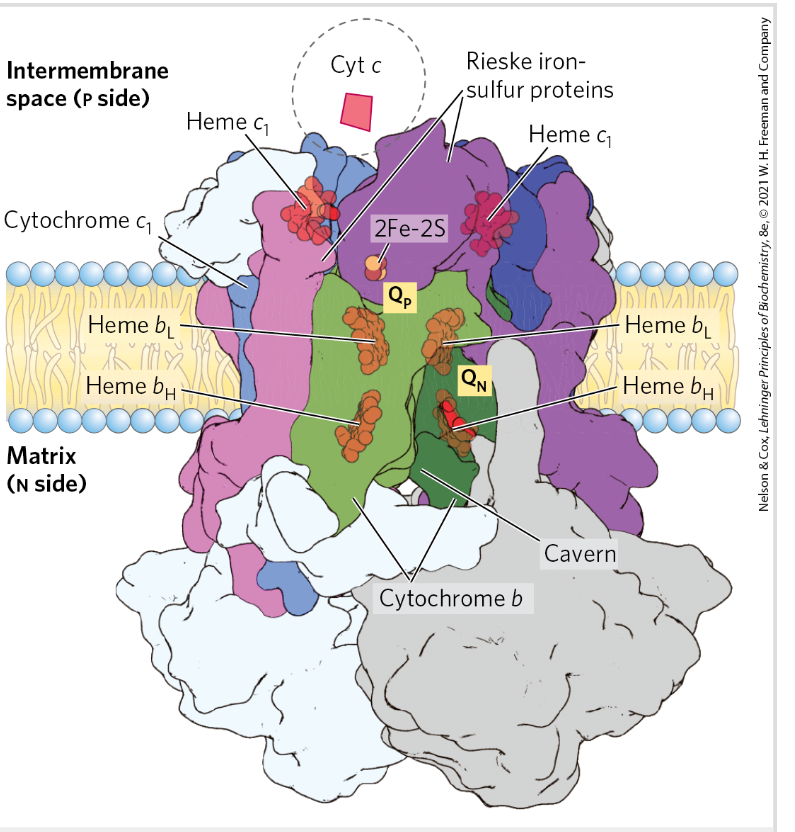
Cyt c
Small soluble problem
Heme group accepts e from complex 3→ moves to the intermembrane space to complex 4 to donate e to a binuclear Cu center
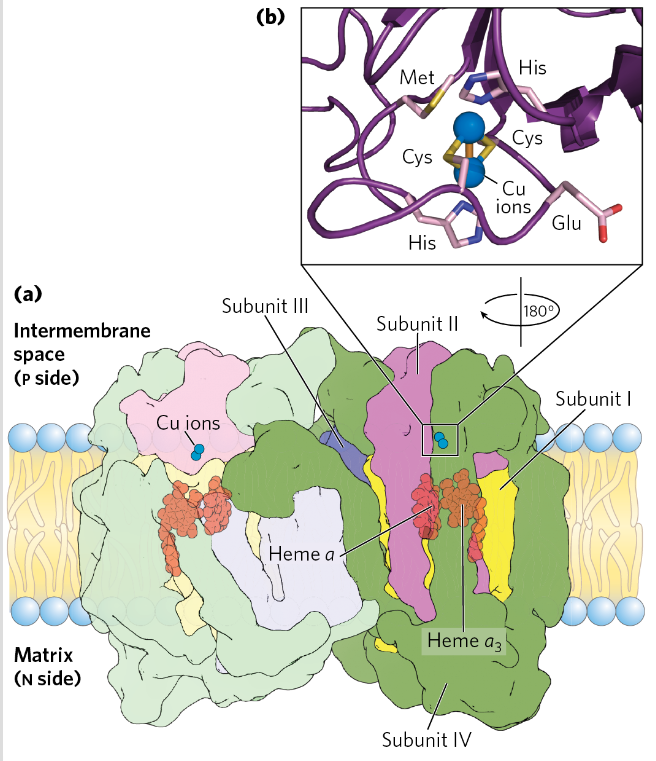
Complex 4= Cytochrome oxidase
Transfers e from cyt c to O2→ H2O
2 Cu ions with SH groups of 2 Cys residues
Pump protons
Heme groups
Cyt C - > CuA→ heme a→ heme a3→ CuB→ O2
Overall reaction: 4 cyt (red) + 8H+N + O2 → 4 cyt c (ox) + 4H+P + 2H2O
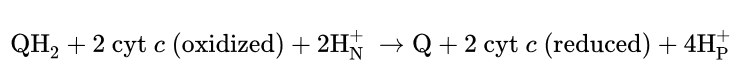
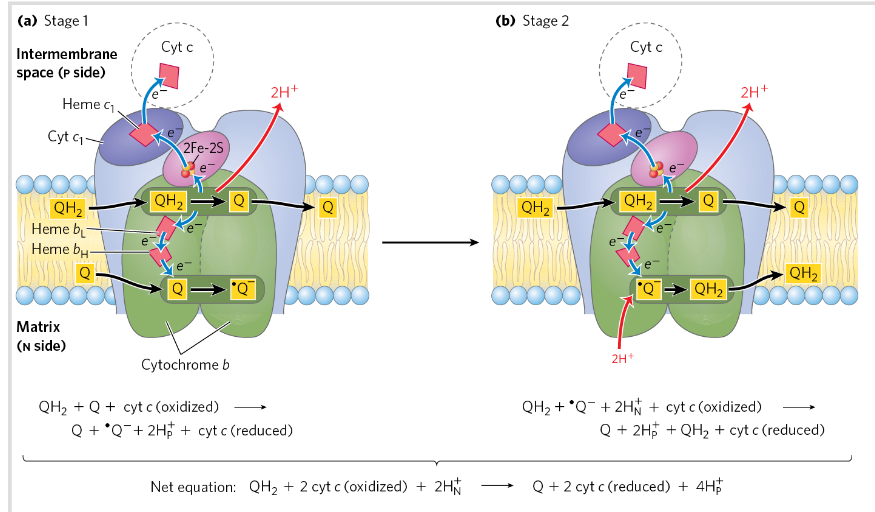
Q cycle
Q cycle is a process occurs in complex 3→ helps transfer e from QH2 (2xe) to cyt c (only carry 1 e)
QH2→ carries 2 e→ Q cycle splits the e to different paths→ efficient transfer
Never a release of the radical formed!
Stage 1: E moves from QH2 (ox) → complex 3 + release H+ at one side
Stage 2: Other side→ Q is reduced and protons are taken up
O2→ H2O
Final electron acceptor
4 e + 4 H+→ heme a3 and Cu can only take one e at a time→ O2 is reduced stepwise to water → intermediate forms like O2- or H2O2
Risk of escaping and forming ROS
Respirasome
Copmplex 1,3,4
Complex 2- free floating in the membrane
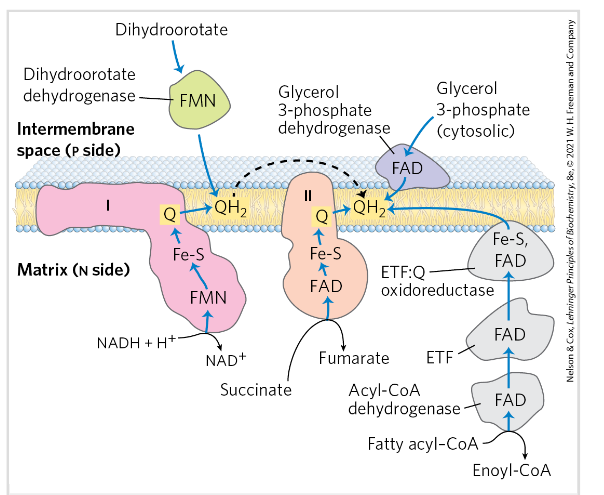
3 enzymes that reduce Q to QH2 in the inner mitochondrial membrane
Acyl-CoA dehydrogenase via ETF (electron transferring flavoprotein) and ETF:Q oxidoreductase (from β-oxidation),
Glycerol 3-phosphate dehydrogenase (from glycerol metabolism or glycolysis),
And dihydroorotate dehydrogenase (from pyrimidine synthesis).
Proton motive force
=PMF
=concentration gradient (due to H+) + electrical gradient ( separation of charge)
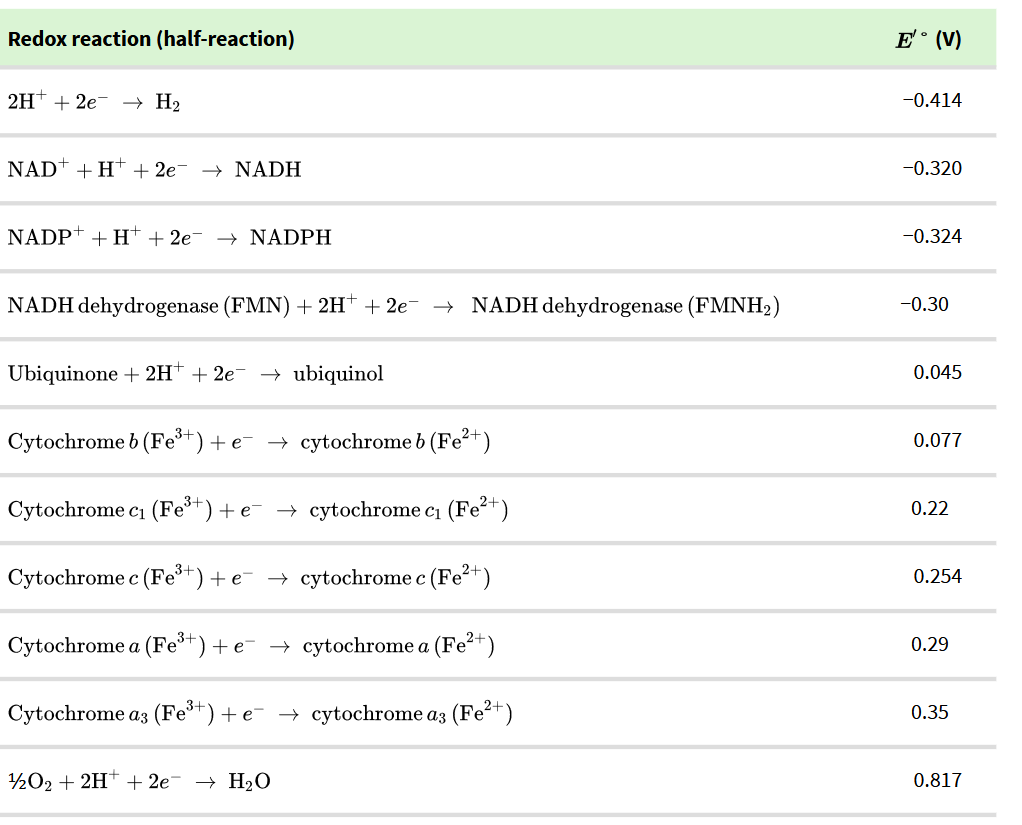
Equation 2 e- through the respiratory chain
2 NADH + 2H+ + O2 → NAD+ + 2H2O
Highly exergonic
ROS
Superoxide
Hydrogen peroxide
Hydroxyl radicals
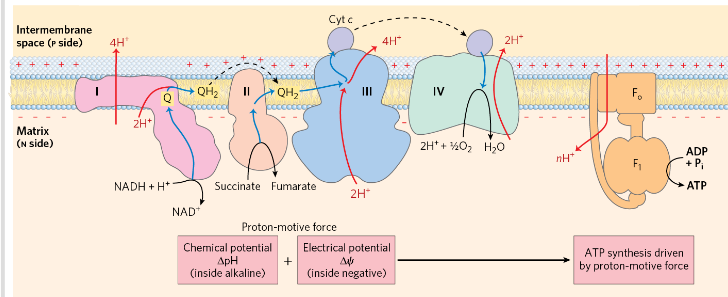
Energy coupling
PMF drives the synthesis of ATP as protons flow passively back into
the matrix through a proton pore in ATP synthase
Coupling= obligate connection between ATP synthesis and e flow through respiratory chain - neither can proceed without the other
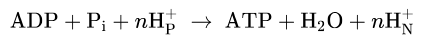
Coupling process
ADP + Pi + succinate →
substrate is oxidized to fumarate
O2 is consumed
ATP is synthesized
Ozygen consumption and ATP synthesis depend on the presence of the substrate! + ADP + Pi
Block ATP synthase
Inhibitors blocking e transfer→ block ATP synthase
Ex oligomycin- block proton channel of ATP synthase→ e transfer stops
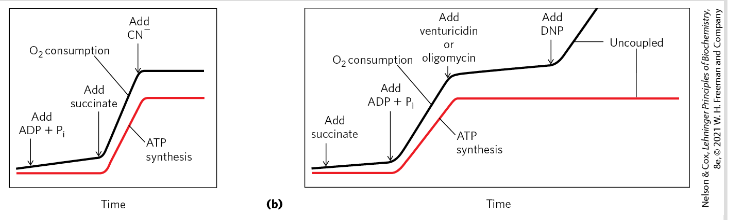
Uncoupling through phosphorylation
Uncouple oxidation from phosphorylation
Still catalyze e transfer from succinate to NADH + O2 but no ATP synthesis
Example compounds: DNP, FCCP
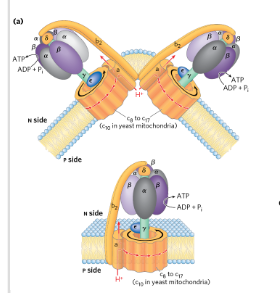
ATP synthase
F type ATPase
ADP + Pi → ATP driven by flow of protons from P→ N
2 components: F0 (into membrane, proton pore) and F1 (peripheral membrane protein, make ATP)
How does the enzyme compensate for the unfavorable energy of ADP-Pi→ ATP
The binding energy of enzyme ATP complex compensate for the unfavorable energy of the reaction
ATP synthase stabilizes ATP relative to ADP + Pi by binding ATP more tightly releasing enough energy to counterbalance the cost of making ATP
Binds ATP with very high affinity and ADP with low affinity→ this drives the reaction to produce ATP
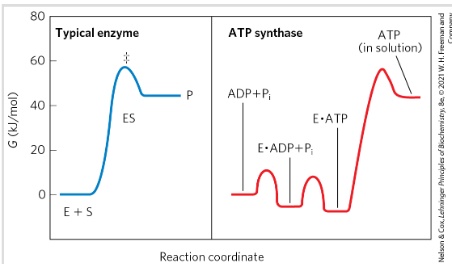
DIfference between typical enzyme and ATP synthase
Typical E: catalyze by lowering activation energy
ATP synthase: Uses a proton gradient to rotate parts of its structure→ mechanical energy converted to chemical energy y synthesizing ATP. Major energy barrier is the release of ATP from E not the reaction
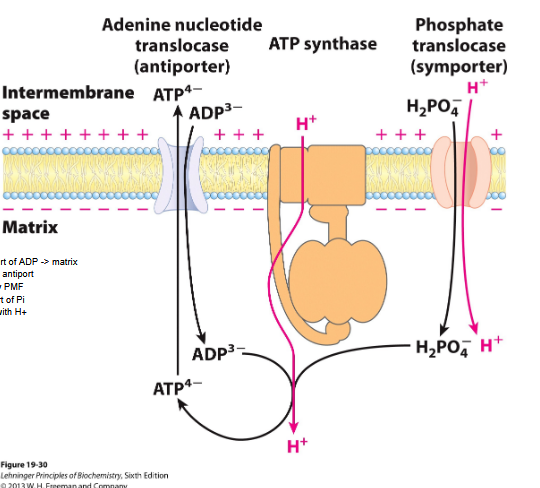
Transport of ADP
) Transport of ADP -> matrix
- ATP/ADP antiport
*) driven by PMF
*) Transport of Pi
- symport with H+
Inner mitochondria mm
Adenine nucleotide translocase binds ADP3—> transports into matrix and exchange for ATP4- transported out
The proton motive force drives ATP-ADP exchange
Phophate translocase- promotes symport of one H2PO4 and one H+
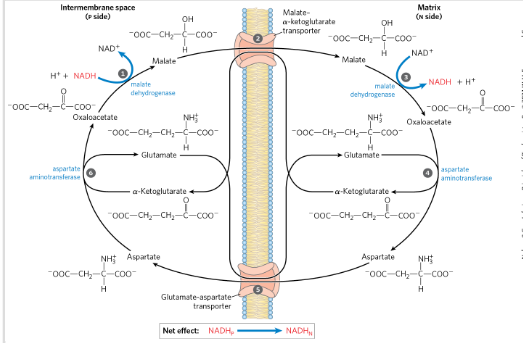
Transport of NADH formed in cytosol to matrix in mito
Through the malate-ASP shuttle
“Loss less”
NADH reduces OAA→ Malate
Malate crosses inner mitochondrial membrane via transporter
Malate is oxidized back to OAA→ NADH in the matrix
NADH to ETC
OAA cant cross the membrane
The shuttle doesnt transport NADH directly but it electrons
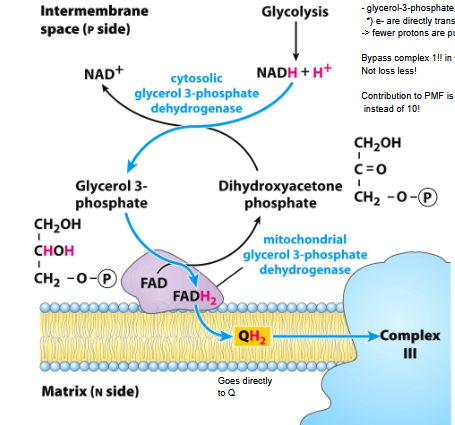
Glycerol-3-phosphate shuttle
Transport in skeletal muscle and brain
) Transport of e from NADH formed i cytosol
) e- are directly transferred to QH2
-> fewer protons are pumped across the inner mm
Bypass complex 1!! in this pathway
Not loss less!
Contribution to PMF is smaller! only 6 protons
instead of 10!
Summary oxidative phosphorylation
SUMMARY:
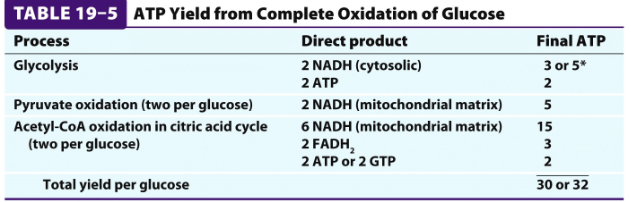
*) 30-32 ATP per glucose -> 976 kJ/mol -> 34% efficiency! Is it good enough? The rest of energy goes of as heat!
*) Thermogenin (UCP1)
*) Thermogenin (UCP1)
- "brown fat"- contain a high density of mitochondria
Contain a lot of heme-> color
Infants has more brown fat-> to keep warm
Regulation of oxidative phosphorylation
- [ADP] is high -> electron transport is very active HIGH
- If [ATP] is high-> e transport goes DOWN
![<p>- [ADP] is high -> electron transport is very active HIGH<br>- If [ATP] is high-> e transport goes DOWN</p>](https://knowt-user-attachments.s3.amazonaws.com/10c96afa-3ffc-4991-b93d-625433520443.png)