final - intro to molecular diagnostics (cls 605)
1/93
Earn XP
Description and Tags
DNA sequencing; amplification; hybridization techniques; etc
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
94 Terms
DNA sequencing (definition)
Methods used to determine the order of nucleotides in a DNA molecular
"gold standard" for mutation analysis
first generation DNA sequencing (general)
In the 70's, two ways to sequence DNA were developed
Maxam Gilbert sequencing
Uses a series of chemicals that cut DNA into fragments at certain bases
Sanger sequencing
Chain termination method using dideoxynucleotide chemistry
more popular method, still heavily used today
basic steps of sanger sequencing
extract DNA
denature DNA
anneal primer to template DNA
synthesis and incorporation of deoxynucleotides and fluorescently labeled dideoxynucleotides (chain termination)
steps 2-4 repeated 30-40 times
detection and sizing of fragments
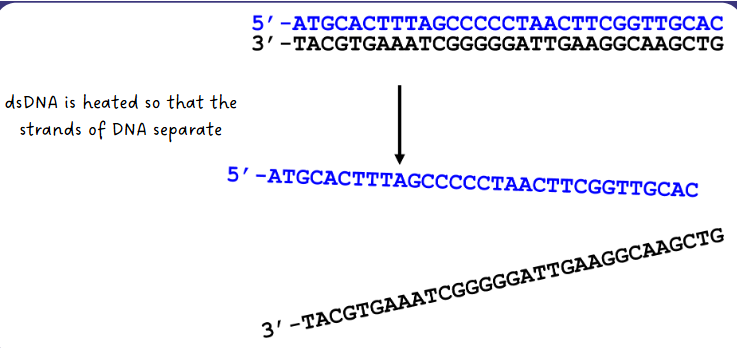
(sanger sequencing) DNA denaturing step
dsDNA is heated so that the strands of DNA separate
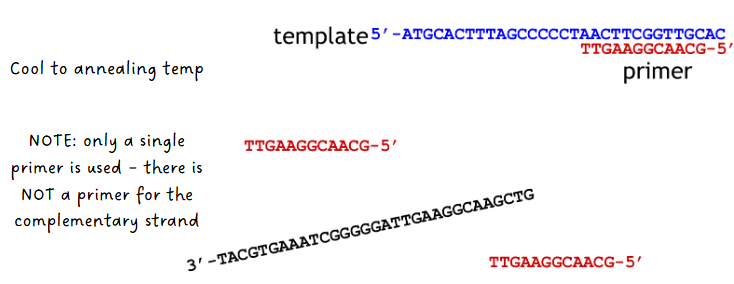
(sanger sequencing) annealing of primer to template step
system cooled to annealing temp
only a single primer is used—there is NOT a primer for the complementary strand
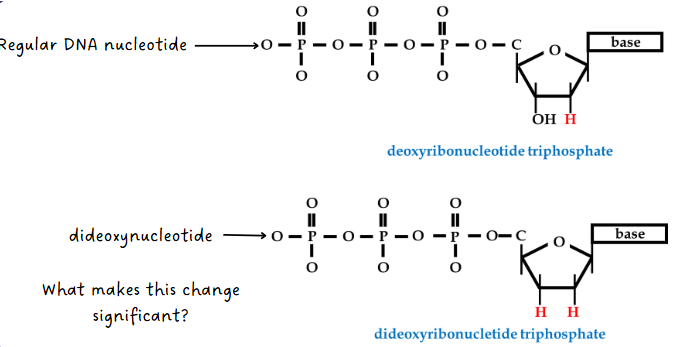
(sanger sequencing) synthesis of new DNA step
The following things are added:
DNA polymerase
dCTP, dTTP, dGTP, dATP (regular DNA nucleotides)
small amounts of ddCTP, ddTTP, ddGTP, ddATP
DNA pol synthesizes complementary strand using the dNTPs but occasionally adds a ddNTP instead which causes chain termination (ddNTP has no 3’ OH but fluoresces when added)

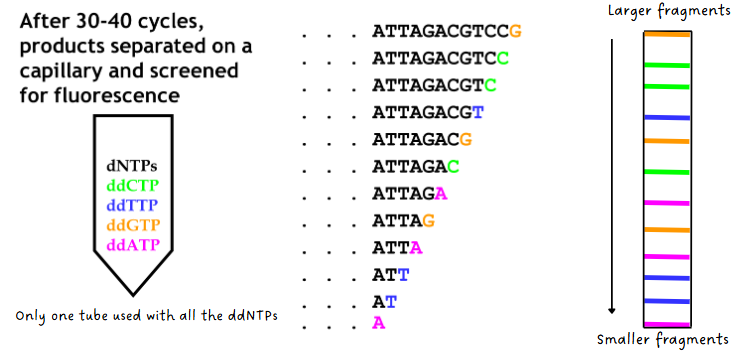
(sanger sequencing) detection & sizing of fragments
old method used radioactive tags instead of fluorescent tags which required 4 separate reaction tubes
new method: after 30-40 cycles, products separate on a capillary and screened for fluorescence (see both pics)
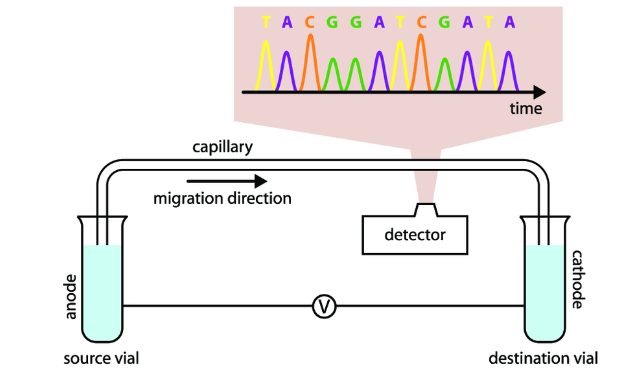
(sanger sequencing) sequence alignment
way of arranging the sequences of DNA to identify regions of similarity that may be a consequence of functional, structural or evolutionary relationships between the sequences
BLAST (basic local alignment search tool) compares an input sequence with sequence in a selected database
limitations of sanger sequencing
can only sequence short pieces of DNA--it doesn't work well for sequences longer than 1000 base pairs
quality is often not very good in the first 15-40 bases because that’s where the primer binds
Sequence quality degrades after 700-900 bases
Low throughput & limited read length
Not accurate in GC-rich and repetitive regions
Relatively fast and cheap for short sequences but inefficient and expensive for long sequences
next-generation (2nd) sequencing
aka massively parallel sequencing
Sequencing large numbers of templates carrying millions of bases simultaneously
Several high-throughput approaches to DNA sequencing
Pyrosequencing
Reversible dye terminator
Ion conductance
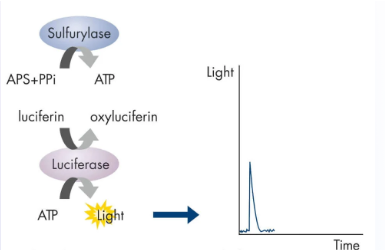
(next gen sequencing methods) pyrosequencing
Detects incorporation of nucleotides during DNA synthesis by monitoring light emitted from a chemiluminescent reaction
Sequencing by synthesis
Reaction mix: ssDNA template, primer, sulfurylase, luciferase, adenosine 5' phosphosulfate (APS), luciferin
example: Roche 454
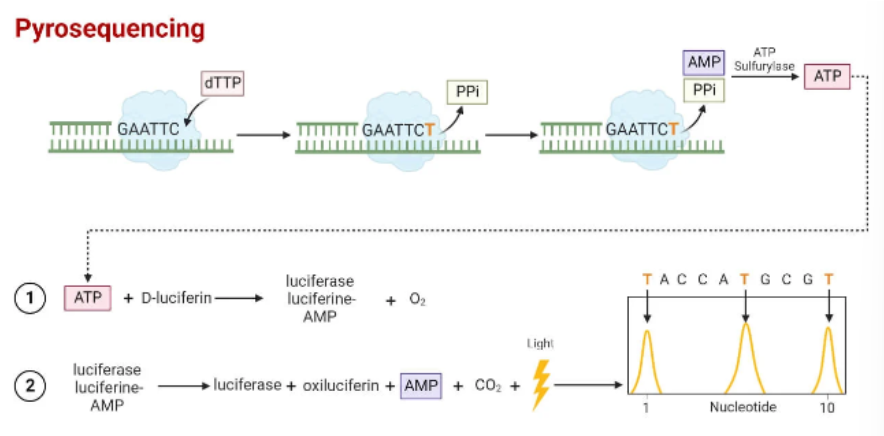
general steps of pyrosequencing (2nd gen)
Sample preparation—DNA extracted and fragmented
PCR amplification—DNA Is amplified, one strand is labeled
Sequencing reaction—light is generated when base is incorporated
Sequence analysis—light signals are analyzed to determine sequence
chemical principle behind pyrosequencing
dNTPs added one at a time—if the nucleotide is complementary to template at next base, DNA polymerase extends primer
Pyrophosphate (PPi) is released when phosphodiester bond forms
PPi converted to ATP by sulfurylase
Luciferase converts luciferin → oxyluciferin
Light released
If nucleotide was not incorporated, it gets degraded by apyrase
Process is repeated
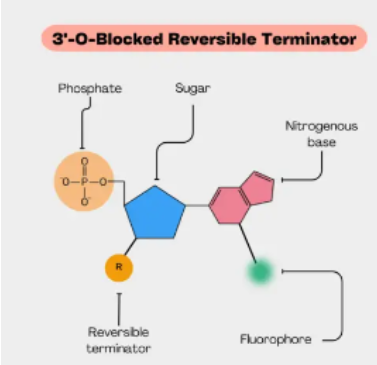
(next gen sequencing methods) reversible dye terminator
Uses fluorescently labeled nucleotides with reversible blocking group
Sequencing-by-synthesis
General steps
Sample prep
Immobilization and amplification
Sequencing
Data analysis
(reversible dye terminator method) Illumina MiSeq
1 million to 25 million reads per run
Run time: 4-56 hours
Output: 540 mb-15 Gb
Lots of short reads
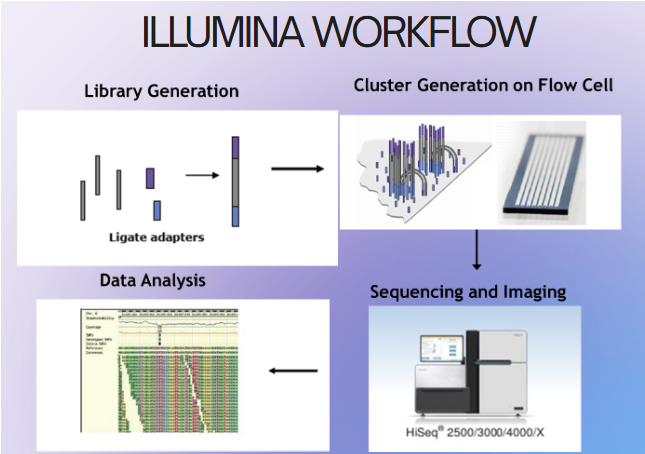
(reversible dye terminator method) illumina library generation
Fragmentation of DNA
End repair of fragmented DNA
Ligation of adapter sequences
Optional library amplification
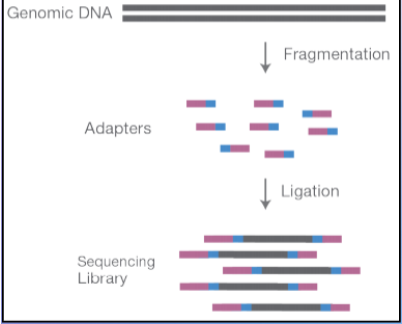
(reversible dye terminator method) adaptors & cluster generation
adaptors: short synthetic DNA sequences added to the DNA fragments
Allow attachment of DNA to flow cell
Priming sites for amplification
Sample indexing (like a barcode)--can run multiple DNA at once
cluster generation: clonal copies of forward and reverse strands in a cluster (500-2000 copies)
increases the signal
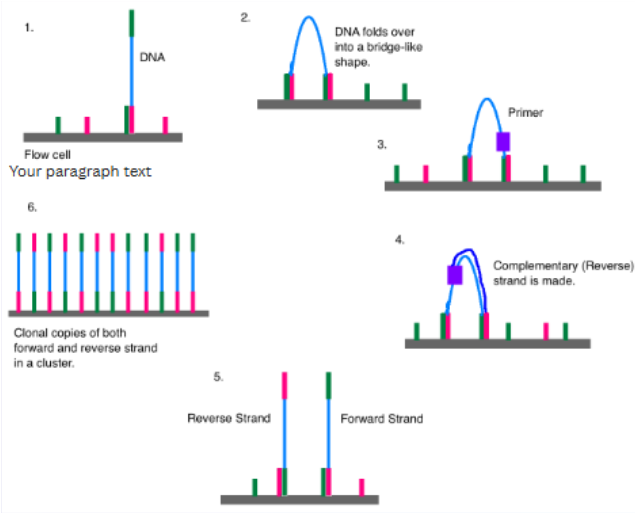
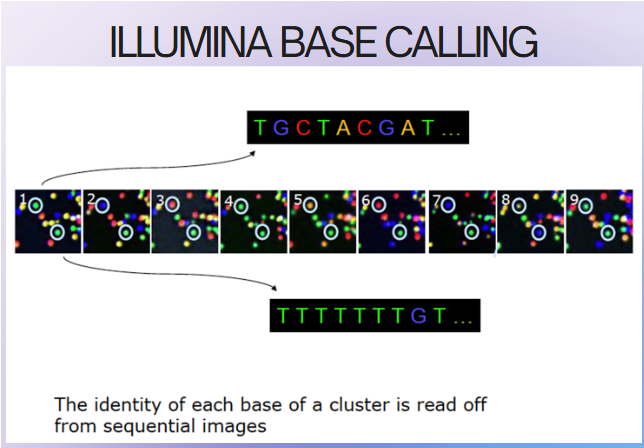
(reversible dye terminator method) illumina sequencing steps
DNA polymerase, connector primers and 4 dNTP w/ base-specific fluorescent markers added to reaction system—3'-OH of these dNTP are blocked, which ensures that only one base will be added at a time
Add bases
1 base gets incorporated
Remove unincorporated bases
Detect signal
Unblock
Repeat
high sensitivity camera captures fluorescent signal emitted by the incorporated nucleotide, identifying the base
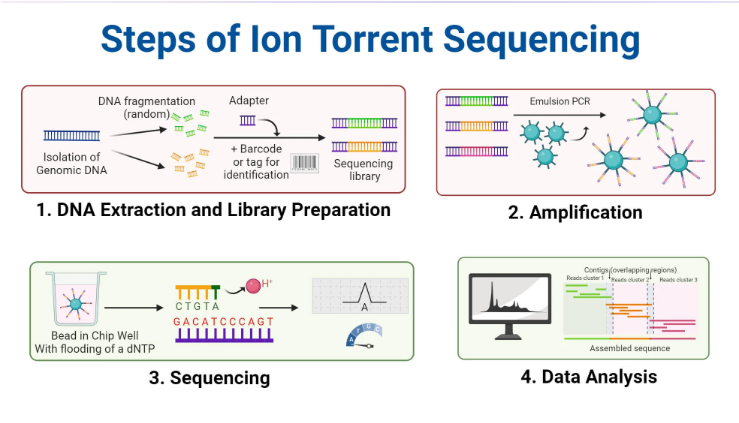
(next gen sequencing methods) ion torrent sequencing
Does not use measure light or fluorescence
Uses semiconductor chips to detect when hydrogen atoms are released during DNA synthesis (pH changes)
steps of ion torrent sequencing (2nd gen)
Beads with amplified DNA are loaded into wells on semiconductor
One at a time nucleotides are flooded across the chip
If that nucleotide gets incorporated, hydrogen ions are released
Ion sensor detects change in pH and converts to electrical signal
(next gen sequencing) read depth
the number of times a specific nucleotide is sequenced
10x read depth means that each nucleotide was sequenced 10 times
Helps with alignment, accuracy and reliability of the results
Increases the confidence that the sequencing is working and that the bases added are accurate
3rd generation DNA sequencing
main difference from 2nd gen: can take longer reads
instruments/technologies: oxford nanopore & pacbio
(3rd gen sequencing) oxford nanpore
Uses engineered proteins to physically ID bases
Nanopore made from the protein alpha hemolysin
Cyclodextrin is localized within the nanopore
Cyclodextrin transiently binds with molecules as they pass through the nanopore
oxford nanpore steps
lipid bilayer created over a microwell containing a pair of electrodes on both sides
nanopores introduced into the bilayer, creating holes
lipid bilayer has a high electrical resistance so current flows only through the nanopore
DNA sample is introduced into the top layer
electrical field pulls charged particles through the pore; particles transiently bind to the cyclodextrin and produce a change in impedance proportional to the volume of the particle
bases are detected and identified by their characteristic impedance change
(3rd gen sequencing) pacbio general info
1 million to 25 million reads per run
Run time: 12-30 hours
Output: 120-480 Gb
Long reads
steps of the pacbio (3rd gen)
Involves a single stranded molecular of DNA, bound to a DNA polymerase enzyme
The bound pair enter a sequencing chamber, called a flow cell
Like in Sanger sequencing, the DNA polymerase adds complementary, fluorescently labelled bases to the DNA strand
As each labelled base is added, the fluorescent color of the base is recorded before the fluorescent label is cut off, the next base in the DNA chain can then be added and recorded

target vs signal amplifcation
Target amplification: increases the number of target molecules
Signal amplification: increases the signal generated by a fixed amount of target
signal amplification methods (4)
Branched DNA (b-DNA)
Hybrid capture
Cleavage-based amplification (invader)
Cycling probes
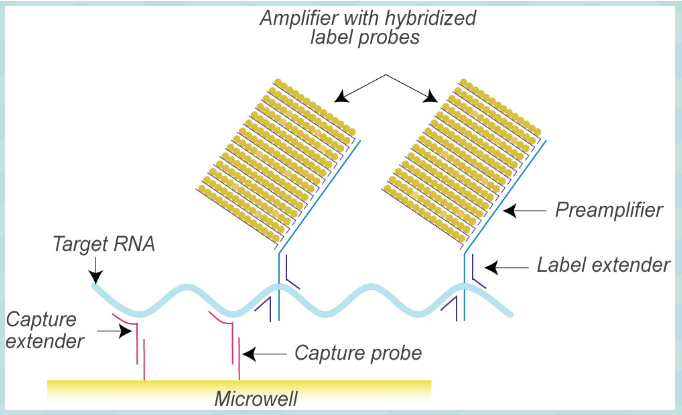
(signal amplification methods) branched DNA (b-DNA)
Short oligo probe captures target nucleic acid onto solid surface
Extender probes bind to target
Multiple reporter probes bind to extender probes--reporter probes bound to alkaline phosphatase labeled nucleotides
Dioxetane (alk phos substrate) added, reaction produces light
branched DNA advantages over PCR + applications
Advantages:
Fast
No need for thermocycler--can do at one temperature
Lower contamination risk--doesn't make a ton of copies that are now floating around
Less chance for nonspecific binding (more specific)
Applications:
Hepatitis B, C testing
HIV testing
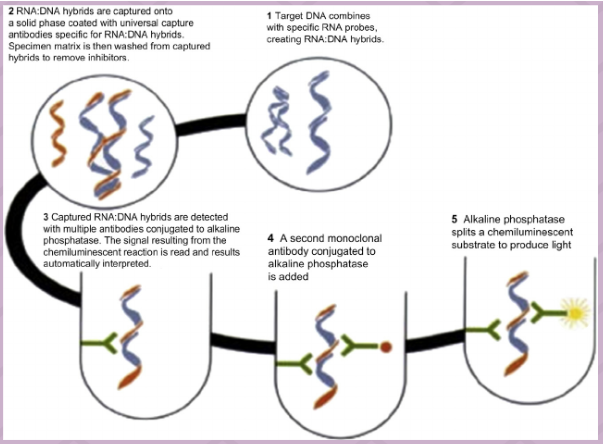
(signal amplification methods) hybrid capture
Target DNA binds to ssRNA probe
DNA;RNA hybrid recognized by antibodies
Antibodies attached to solid surface and capture DNA;RNA hybrid
Secondary antibodies attached to alkaline phosphatase added (sandwich assay)
Substrate for alkaline phosphatase added--light produced
Applications: HPV & genitourinary specimens, hepatitis viruses, CMV; also a hybridization technique
(signal amplification methods) cleavage-based/invader amplification
Overlapping probes bind to target sequence (invader probe, signal probe)
Cleavase cuts in overlapping region--cleaves signal probe
FRET probe added (intact probe produces no signal) that has complementarity to the cut part of the signal probe
Signal probe (now an invader probe) binds to FRET probe--due to folding of FRET probe this makes a three strand overlap
Cleavase recognizes overlap and cleaves
Signal gathered from FRET probe
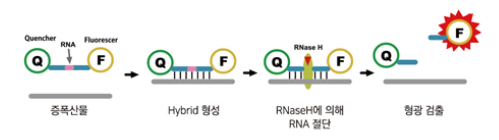
(signal amplification methods) cycling probes
probes made of DNA and RNA with a reporter at one end and a quencher at the other
If complementary sequence is found, probe will anneal
RNAse H cleaves RNA part of probe; releases reporter from quencher
Applications:
Detect genes associated w antimicrobial resistance (vanA, mecA)
Detection of pathogens (herpes virus)
target based amplification methods
usually known as isothermal amplification
done at constant temp & does not involve temperature changes
examples: transcription mediated amplification and nucleic acid sequence based amplification (TMA and NASBA), loop mediated isothermal amplification (LAMP)
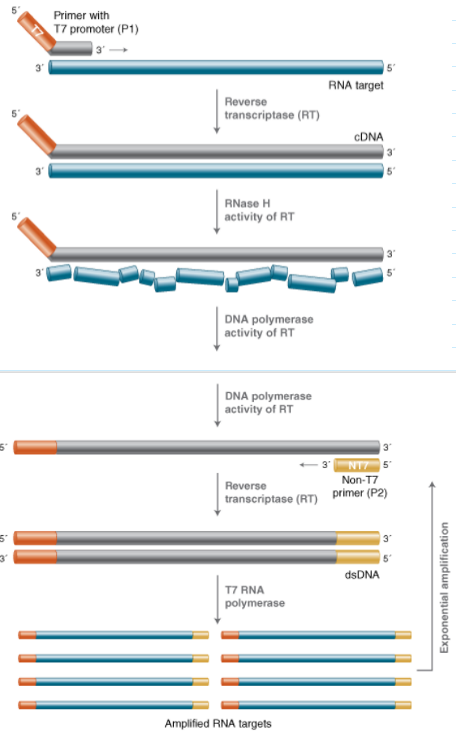
(target/isothermal amplification) transcription mediated amplification
Uses amplification through RNA instead of DNA
Target molecule can be RNA or DNA
Primers are designed to target a region of interest, but one primer includes the promoter sequence for T7 RNA polymerase at the 5' end
If RNA Is the target: RNA → cDNA → RNA
Benefits: RNA sensitivity prevents possible contamination
Drawbacks: RNA is more sensitive to degradation
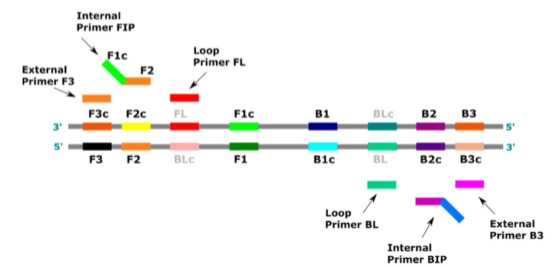
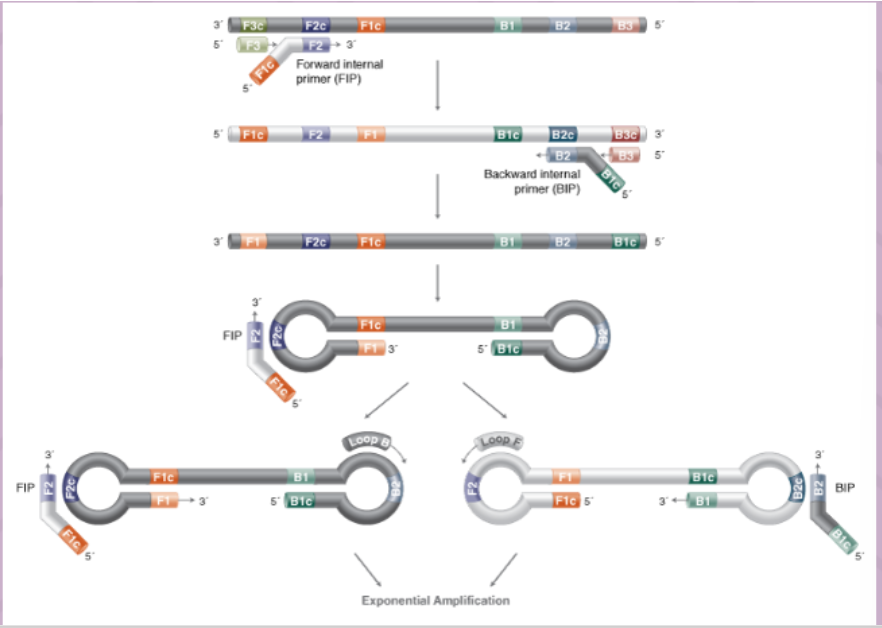
(target/isothermal amplification) loop mediated amplification (LAMP)
uses 4-6 primers recognizing 6-8 distinct regions of target DNA
A strand-displacing DNA polymerase initiates synthesis and 2 specifically designed primers form "loop" structures to facilitate subsequent rounds of amplification thru extension on the loops and additional annealing of primers
DNA products are very long (>20 kb) and formed from numerous repeats of the short (80-250 bp) target sequences
can produce copies without need for denaturing the DNA
probe amplification methods
Instead of increasing the amount of target molecule some methods increase the amount of probe
Examples: Q-beta replicase, ligase chain reaction
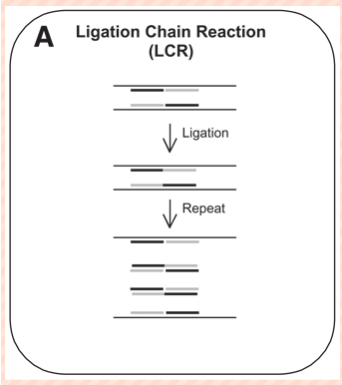
(probe amplification) ligase chain reaction
dsDNA denatured by heat → single-stranded target sequence
temp lowered and 2 specific probes anneal to amplicon
Another 2 probes bind to the amplicon's complementary sequence on the other strand
When two probes anneal correctly to a target sequence, the 5' end of one probe is next to the 3' end of the other
The probes can then be covalently joined by a heat-stable ligase to form a new target sequence--to which another pair of probes can subsequently bind
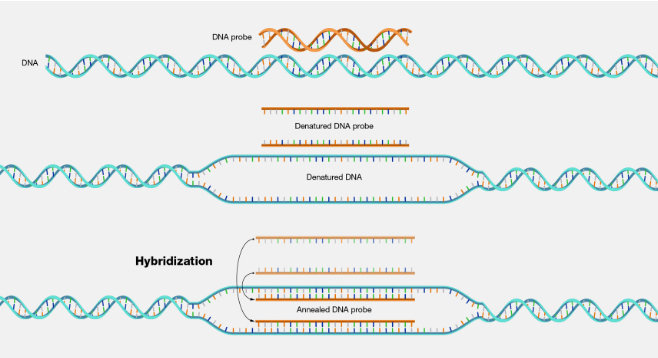
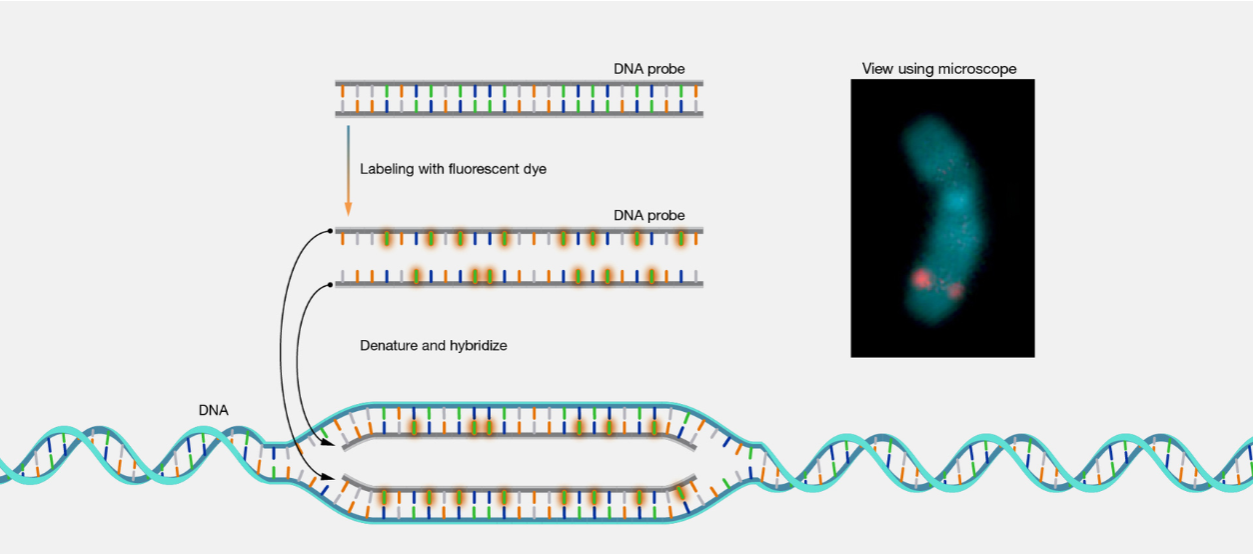
hybridization (definition)
annealing (base-specific hydrogen bonding) of a probe to a target nucleic acid to form a double stranded molecule
probe: typically a single stranded DNA, RNA or PNA molecule, about 20-2000 bases in length
target: ssDNA or RNA molecule
applications of hybridzation techniques
Because DNA probes with a known sequence will anneal to targets based on complementation, information can be gained about the sequence of the target DNA
e.g. the presence or absence of mutations
applications:
Southern blots (DNA is the target)
Northern blots (RNA Is the target)
In situ hybridization
Dot blots
Hybrid capture
Microarrays
western blot looks for _____ using _____?
looks for proteins using antibodies
northern blot looks for _____ using _____?
looks for RNA using RNA/DNA probe
southern blot looks for _____ using _____?
looks for DNA using RNA/DNA probe
(hybridization) probes
single stranded DNA, RNA, or PNA molecule about 20-2000 bases in length
Complementary to target
Usually labeled in some way
e.g. fluorescent molecule, ligand, isotope
Common source of probes
Synthetic oligonucleotide
PCR product
Restriction endonuclease fragment
hybridization principle (pic)
southern blot
Develop in 1975 by Edwin Southern
Purpose: determines if there is a certain DNA sequence in a sample
General steps:
Extract DNA
Cut DNA with restriction enzymes
Gel electrophoresis
Blotting (transfer)
Probe hybridization
Detection
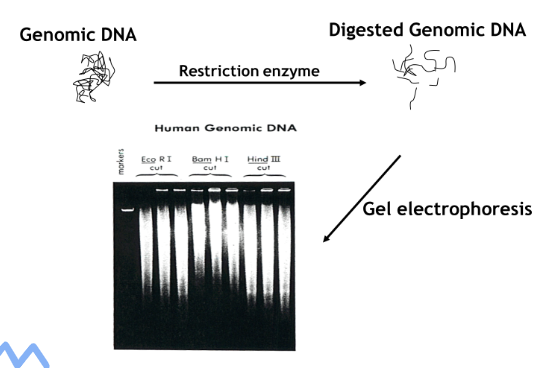
(southern blot) restriction enzyme cutting and resolution
Enzymes used depends on gene of interest and application
Digestion for extended amount of time to allow complete cutting
Run gel electrophoresis with ethidium bromide
Digested genomic DNA should look like a smear
Problems:
Large aggregate of DNA at top = uncut
Smear mostly at bottom = degraded DNA
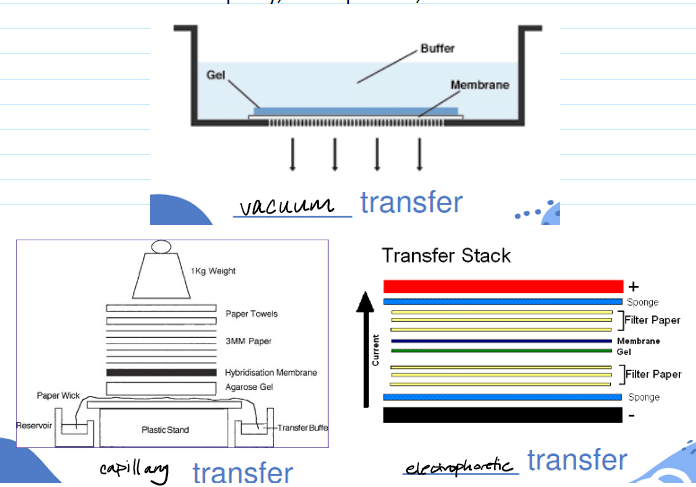
(southern blot) blotting/transfer step
DNA transferred to solid surface so that you can then hybridize with the probe
Possible substrates: nitrocellulose, nylon, modified cellulose, others
Transfer methods: capillary (longest but most simple), electrophoretic, vacuum
(southern blot) pre-hybridization step
After transfer, DNA may be permanently immobilized by baking or UV cross-linking
Prevents DNA from moving or washing away
Pre-hybridization wash or blocking solution is added to prevent probe from non-specifically sticking to membrane
(southern blot) hybridization step
Complementary to target gene
Complementary sequences are not identical--they are antiparallel
Example: if this is our target sequence (pic), what is probe sequence?
Answer: 5' ATCAGCGAGCTAC 3'
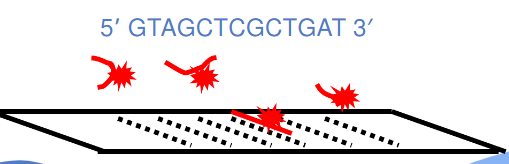
(southern blot) detecting probes (pic)
southern blot application—RFLP
Differences in DNA sequences between/within individuals can result in variation in the position or cleavability of restriction endonuclease (RE) site
may be the result of mutations (and other changes) associated with specific genetic diseases/conditions
These variations may be detected by examining the pattern or chromosomal RE fragments
(southern blot) RFLP applications
Genetic disease/condition diagnosis
A standard RFLP can be used in diseases:
Involving point mutations resulting in the loss (or gain) or restriction enzyme site
Deletion or insertion mutations involving at least 50 base pairs
Associated with altered DNA methylation
Genetic identity testing
(RFLP applications) fragile X
most common cause of inherited intellectual disability
Symptoms: physical and behavioral features and delays in speech and language development
Caused by mutation of the FMR1 gene which codes for FMRP (fragile X messenger ribonucleoprotein)
FMRP is most actively synthesized in the brain
Females are less affected because they have two X chromosomes (and hence 2 copies of the FMR1 gene)
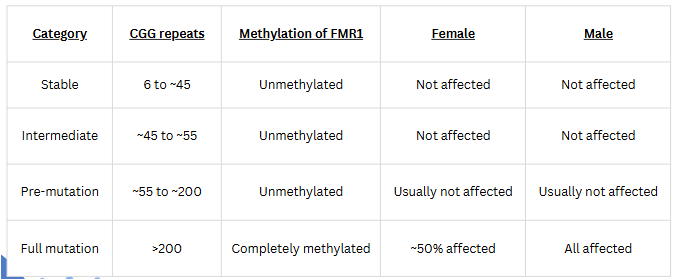
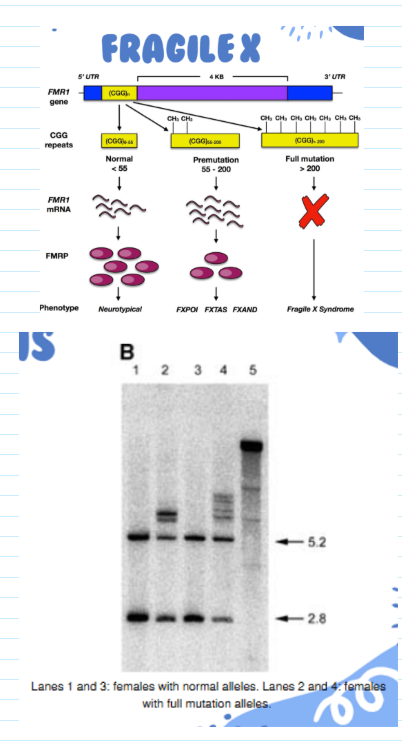
molecular diagnosis of fragile X
Extensive repeat extension (>200) leads to methylation of C residues in CpG islands
RFLP results from loss of cutting by methylation-sensitive restriction enzymes (e.g. Xhol)
RFLP detected by hybridization with FMR probe
Current diagnostic testing:
PCR as first line test
Southern blot (RFLP) as follow up
Why not use PCR only?
PCR doesn't give any information on methylation status and that is needed for diagnosis/severity
other blots/modifications of southern blot procedure
Northern blot--used to detect RNA
Western blots--uses antibodies to detect proteins
HIV confirmatory testing, lyme disease, variant Creutzfeldt-Jakob Disease (vCLD)
dot blots
microarrays
(hybridization) dot blots
Simplified version of Southern or Western blotting
Sample applied directly to membrane without electrophoresis first
Presence and amount of target detected by use of labeled probes
What does a darker dot mean?
more of whatever you are detecting (semi-quantitative)

(hybridization assay) Digene HC2 HPV DNA Test
Screens for presence or absence of 13 types of high-risk HPV that are most associated with cervical cancer
negative HPV test and normal Pap result provide confidence that a woman does not have, and is not likely to develop, high-grade cervical disease or cancer within the next 3 years (99.21% NPV)
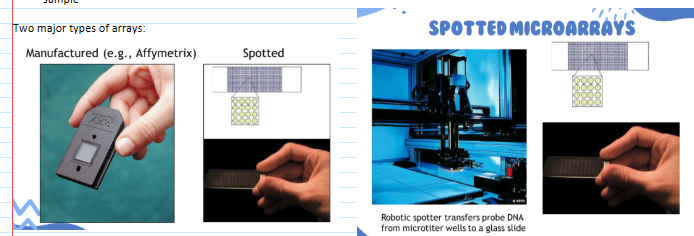
(hybridization) microarrays
A technology that can simultaneously detect thousands of specific DNA sequences in a short TAT
two types: manufactured & spotted
Applications:
Oncology: analyze gene expression in cancer cells
Infectious disease: detect the presence of nucleic acids from disease-causing agents
Mutation analysis: detect single nucleotide changes to large insertions or deletions
principle of microarrays
Multiple probes are spotted to a solid surface (microscope slide or silicone chip)
Hundreds to hundreds of thousands of different probes—may be representation of the entire genome, specific parts of a genome, infectious agents etc
Fluorescently-labeled DNA from sample(s) is hybridized to the entire array of probes
Array is scanned with a laser to detect hybridization
The position of fluorescing spots identifies the nucleic acids present in the sample
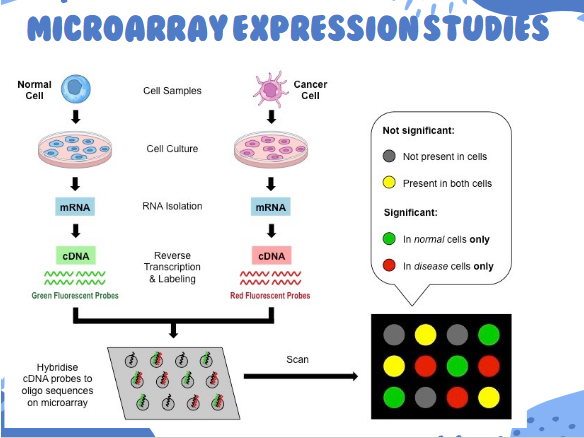
microarray applications
Infectious disease diagnosis
Probes for various pathogens
Gene expression analysis
e.g. diagnose cancer type and treatment, identify genes expressed in response to drugs
Sequence change identification
Identify SNPs associated with genetic disease
Copy number imbalance identification
Array Comparative Genomic Hybridization (arrayCGH)
Identify chromosomal deletions and duplications
in situ hybridization
A technique to detect the presence or absence and location of specific DNA or RNA sequences within preserved chromosome preparations, fixed cells or tissue sections
Can visualize chromosomal deletions, duplications, translocations, copy number variations (ploidy)
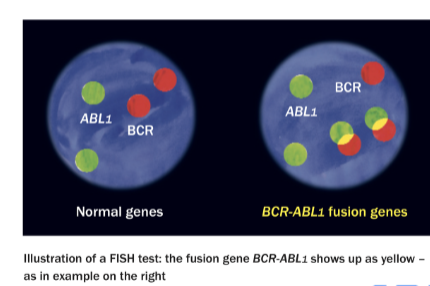
(in situ hybridzation) FISH for BCR/ABL
Translocation between chr 9 and 22 (Philadelphia chromosome) assoc with CML
Detected by hybridization with BCR and ABL probes
(hybridization) stringency
the tolerance for mismatch in base-pairing
If a probe is complementary to a target sequence it will bind
Stable base pairing requires about an 80% or greater match
high vs low stringency
high: conditions where the probe must completely match the target sequence to bind
low: conditions where the probe binds even if it doesn’t match the sequence very well
what happens in high stringency conditions?
detect less of target sequence but also less non-specific binding
what happens in low stringency conditions?
more detection of target, but more non-specific binding
(hybridization) manipulating stringency
Temperature: increase temp increases stringency
Na+ conc: increase conc = decrease stringency
Formamide & urea: increases stringency
"stringency" applies to both the hybridization steps and the subsequent washing steps
reasons for specimen rejection
Incorrect labeling
Not enough sample
Wrong container
Transportation issues
Specimen issues (hemolysis)
Specimens not received in time (esp for RNA)
considerations for infectious disease testing
Type of sample depends on where the pathogen is found, replicates or sheds from
Selection of sample type depends on organism, patient symptoms, and invasiveness of collection procedure
considerations for genetic disease testing
Blood should be an acceptable specimen (i.e. white cells) to detect inherited mutations and to establish identity (paternity, forensics) as would be any cellular material (i.e. buccal smears, hair follicles, semen)
Availability and/or ease of access may determine specimen used
considerations for cancer testing
Malignant diseases usually require the specific tissue affection (ex: biopsy)
However, mutations that make a person more susceptible to cancer (ex. BRCA1) can be detected with any cellular sample (blood)
(specimen requirements) southern blot
Requires (a lot of) high quality DNA
Fresh or quick frozen tissues
If blood is sample type, need 5-10 mLs
(specimen requirements) PCR
Can be done on degraded DNA
Fixed/embedded tissues generally ok
Freezing usually acceptable
(specimen requirements) FISH/ISH
Typically done on intact preserved tissues
Cells don't need to be living
No freezing
(specimen requirements) karyotyping
Need living cells
Whole blood and bone marrow, need anticoagulant
No freezing
(specimen requirements) next gen sequencing
Usually 3-5 mLs of blood
Freezing samples ok
if 100% of the DNA is recovered, how much DNA could be extracted from a 1 mL sample containing 5,000 WBCs/ul?
(5000 WBCs/ 1 uL) x (1000 uL/1 mL) = 5,000,000 cells
5,000,000 cells x (1 ug DNA/151,000 cells) = 33.1 ug DNA
You are extracting DNA for a genomic analysis from a whole blood specimen containing 7,600 WBC/uL. You need 10ug of DNA for your procedure. Assuming you recover 50% of the DNA from the specimen, what is the minimum volume of blood you need for the analysis?
10 ug DNA x (151,000 cells/1 ug DNA) = 1,510,000 cells
1,510,000 cells x (1 uL/7600 cells) = 198.7 uL
198.7/0.5 = 397.4 uL
397.4 = 0.4 mL
(specimen collection; anticoagulants) EDTA
preferred anticoagulant for PCR
EDTA chelates Mg2+, which inhibits nucleases
Not for karyotyping or cell culture
Can be stored at 4 deg C for 72 hours without DNA degradation, need to freeze for longer time periods
(specimen collection; anticoagulants) sodium heparin
NOT FOR PCR
Used for in situ hybridization and karyotyping
(specimen collection; anticoagulants) ACD
Suitable for PCR but will create a dilution that needs to be accounted for
Not acceptable for karyotyping
(sample types) tissue
Total quantity of importance to provide adequate, extractable DNA
tissue quantity should be equal to the size of a pencil eraser ( 3mm x 3mm x 3mm)
In PBS for collection (phosphate buffered saline)
For transport, PBS drained and either dry ice or 70% ETOH used at RT
Storage should be at 4 deg C
Tissues must be received in viable and sterile condition if cell culture is needed (e.g. karyotype)
(sample types) swabs
Swabs for PCR should be synthetic (no cotton or wood)
Flocked swabs enhance sample collection
Mucus should be removed (can be inhibitory)
(sample types) sputum
Can be used for infectious disease testing
Not ideal for other testing due to presence of normal flora and mucus
PCR inhibitors present in samples
Blood—hemoglobin, IgG, lactoferrin
Feces—bile salts
Urine—urea
Sputum—mucus
(transport/storage) RNA
overnight shipping
avoid changes in temperature while shipping
(transport/storage) whole blood/serum/plasma & bone marrow
Whole blood = transport w/in 24 hours, store at RT for 24 hrs, refrigerate up to 72 hours
freezing generally not recommended
Serum/plasma = usually for infectious disease testing
if testing for RNA viruses, spin and freeze immediately, ship on dry ice
Bone marrow = handle like whole blood generally; refrigerate for up to 48 hours
freezing generally not recommended
(transport/storage) tissues & swabs
Tissues = freeze at -20 deg C, transport on dry ice
Swabs = generally transport in sterile saline or UTM (universal transport media), typically refrigerate for up to 72 hours after collection
freeze if specimen can't be processed within that time
storage of extracted nucleic acids
DNA = can store in the refrigerator for up to a week, good for months or years when stored at -20 or -70 deg
RNA = store in refrigerator if using within a few days, store at -20 for use within a few months, store at -80 in liquid nitrogen for longer storage
principle of the biofire film array
nested multiplex PCR
creates melting curve to analyze results
assays: respiratory, pneumonia, blood culture, GI, meningitis/encephalitis panels
principle of diasorin liaison instrument
qualitative, quantitative, and multiplex PCR
no DNA extraction required
assays: COVID-19, Flu A/B, HSV 1 & 2, Bordetella spp., Group A Strep
principle of the hologic panther
proprietary real-time transcription mediated amplification to detect and quantify target sequences
assays: women’s health & infectious disease panels (HIV, HCV, covid etc)
principle of the genexpert instrument
catridge-based nested real-time PCR (qual/quantitative)
numerous assays:
respiratory
infectious disease; TB
virology and sexual health
oncology and human genetics