RNA translation
0.0(0)
0.0(0)
Card Sorting
1/44
Earn XP
Description and Tags
Study Analytics
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
45 Terms
1
New cards
Genetic code
encodes all 20 amino acids
multiple codons for most amino acids
multiple codons for most amino acids
2
New cards
Start codon
AUG start codon
3
New cards
What are the start codons
UAA
UAG
UAG
4
New cards
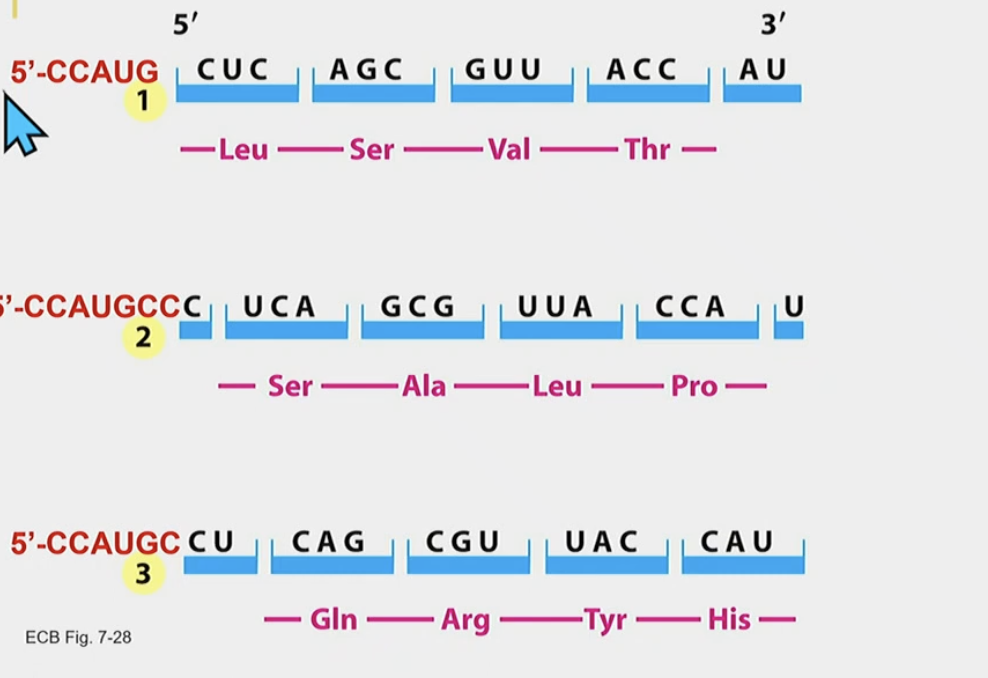
Reading frames…
Defines the amino acid sequence
We define it by looking for the AUG (methionine) sequence and then start grouping the RNA into 3s
We define it by looking for the AUG (methionine) sequence and then start grouping the RNA into 3s
5
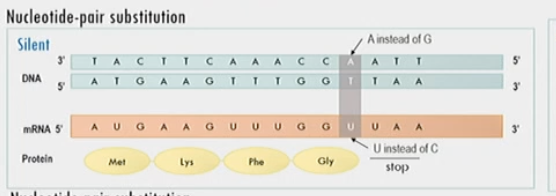
New cards
Silent mutation
nucleotide pair substitution that doesn’t change nucleotide
6
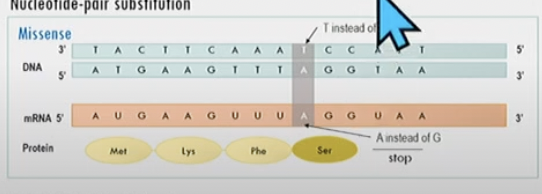
New cards
Missense mutation
nucleotide base-pair substitution
amino acid changesN
amino acid changesN
7
New cards
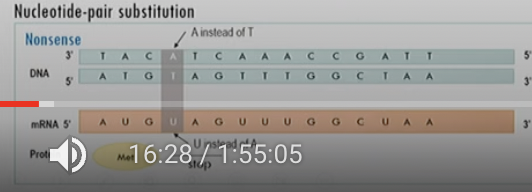
Nonsense mutation
Nucleotide bp substitution
causes a stop codon (UAA or UAG) to appear
causes a stop codon (UAA or UAG) to appear
8
New cards
Nucleotide-pair deletion can result in
Frameshifts..
9
New cards
What can frameshifts result in
can cause immediate missense (amino acid change) or nonsense (stop codon) both, or none!
None happens when a whole amino acid is added or a whole amino acid (3 bps) is deleted
None happens when a whole amino acid is added or a whole amino acid (3 bps) is deleted
10
New cards
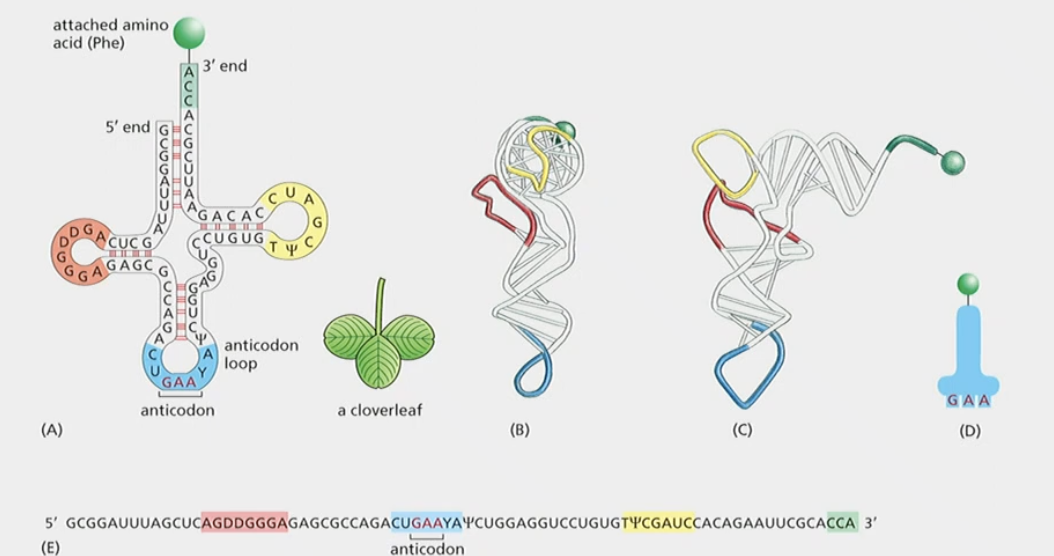
tRNA (4 points)
The link between mRNA and protein sequence
1. recognizes codon on the mRNA and brings in the proper amino acid
2. 80 nucleotides long
3. transcribed from genes as per usual
4. **Base-pairs with itself to form cloverleaf shape**
* amino acid is binded to tis 3’ end
* anticodon, which basepairs with codon antiparallel and complenetary
1. recognizes codon on the mRNA and brings in the proper amino acid
2. 80 nucleotides long
3. transcribed from genes as per usual
4. **Base-pairs with itself to form cloverleaf shape**
* amino acid is binded to tis 3’ end
* anticodon, which basepairs with codon antiparallel and complenetary
11
New cards
How long are tRNAs?
80 nucleotides long
12
New cards
Anticodon
* anticodon, which basepairs with codon on the mRNA antiparallel and complenetary
13
New cards
Dihydrouridine
‘DD’ on left loop
A base that got modified after transcription
A base that got modified after transcription
14
New cards
Pseudoeuridine
Ψ
A base that got modified after transcription
A base that got modified after transcription
15
New cards
Are there Us in DNA?
Yes, sometimes due to mutations
16
New cards
Are there only Gs As Us and Cs in RNA?
No, you have pseudouridine and dihydrouridine that changed after transcription
17
New cards
How codon redundancy is managed for translation
More than one tRNA (different anticodon for different codons of the same amino acid)
but some tRNA have recognize multiple codons due to the **wobble position**
but some tRNA have recognize multiple codons due to the **wobble position**
18
New cards
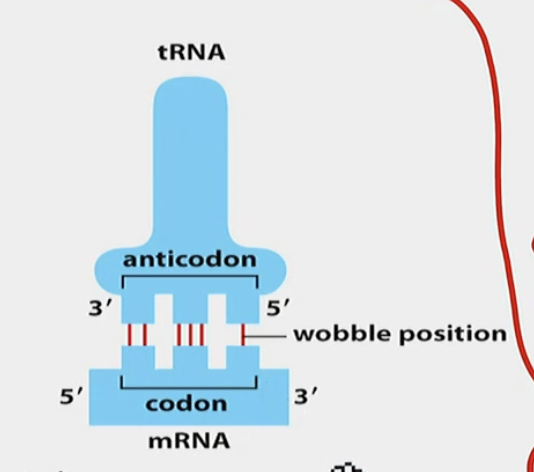
Wobble position
5’ on the anticodon, 3’ on the codon. 3RD POSITION OVERALL.
U can have either A, G, or I.
I is inosine, another modified base
Each base on the codon can have different possible anticodon bases
EUKARYOTES ARE LESS WOBBLY THAN PROKARYOTES
U can have either A, G, or I.
I is inosine, another modified base
Each base on the codon can have different possible anticodon bases
EUKARYOTES ARE LESS WOBBLY THAN PROKARYOTES
19
New cards
2 sequential steps in ensuring fidelity
→ aminoacyl-tRNA synthetases
→ base pairing
→ base pairing
20
New cards
aminoacyl-tRNA synthetases
Proteins that recognize the tRNA and put on the proper amino acid using ATP reaction.
**APPROXIMATELY** 20 synthetases, **1 FOR EACH** AMINO ACID, depends on # of amino acid since not all bacteria have 20
Capable of error correction by **hydrolytic editing**
**APPROXIMATELY** 20 synthetases, **1 FOR EACH** AMINO ACID, depends on # of amino acid since not all bacteria have 20
Capable of error correction by **hydrolytic editing**
21
New cards
Hydrolytic editing
If aminoacyl-tRNA synthetases happen to put the wrong amino acid on the tRNA they can backspace/delete it
22
New cards
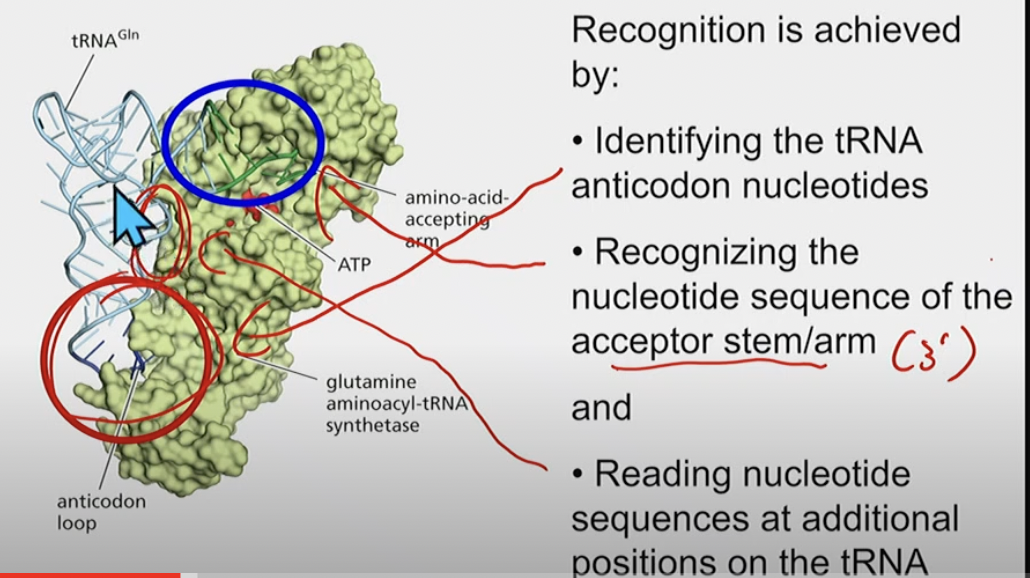
Recognition of a specific tRNA by its synthetase
* identifies (since it has to bind to) anticodon nucleotides
* recognizes the nucleotide sequence of the 3’ acceptor stem/arm (where amino acid is added)
* reading nucleotide sequences at additional positions on the tRNA
* recognizes the nucleotide sequence of the 3’ acceptor stem/arm (where amino acid is added)
* reading nucleotide sequences at additional positions on the tRNA
23
New cards
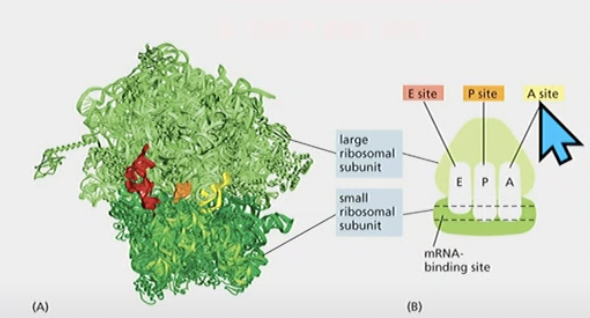
Ribosomal structure
* Made up of different proteins and RNA molecules
* large subunit and small subunit
* 3 important sites, E site P site and A site
* large subunit and small subunit
* 3 important sites, E site P site and A site
24
New cards
Where are ribosomes located
* on endoplasmic reticulum (rough?)
* in cytosol
\
* in cytosol
\
25
New cards
A site P site and E site and what order they appear in
A site = aminoacyl site
* The next tRNA w next amino acid added
P site = peptidyl site
* tRNA with growing polypeptide change
E site = exit site
* where tRNA leaves
* The next tRNA w next amino acid added
P site = peptidyl site
* tRNA with growing polypeptide change
E site = exit site
* where tRNA leaves
26
New cards
Overview of translation
* energy stored in covalent bond btwn amino acid and tRNA in P site makes peptide synthesis energetically favourable (moving peptide bond from P site tRNA to A site tRNA)
* peptidyl transferase activity of the rRNA in the large subunit
\
* peptidyl transferase activity of the rRNA in the large subunit
\
27
New cards
What catalyzes peptide bond formation?
peptidyl transferase activity of the rRNA in the large subunit
28
New cards
Explain a ribozyme
RNA molecules that posess catalytic activity
* therefore, the ribosome is a ribozyme, because **A, P, E sites are primarily made up of RNA molecules**
\
* therefore, the ribosome is a ribozyme, because **A, P, E sites are primarily made up of RNA molecules**
\
29
New cards
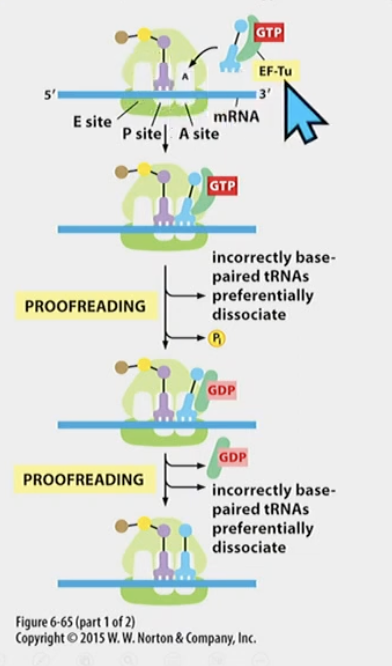
Does the ribosome have a backspace mechanism? How does elongation and quality control happen
Elongation Factors
* EF-Tu in **prokaryotes** (**EF1 in eukaryotes**) checks aminoacyl tRNA
* Ef-G (prokaryotes) (EF2 in eukaryotes)
* EF-Tu in **prokaryotes** (**EF1 in eukaryotes**) checks aminoacyl tRNA
* Ef-G (prokaryotes) (EF2 in eukaryotes)
30
New cards
EF-Tu/EF1
* binds to GTP
* ribose and 3 phosphates
* It checks base-paring. if base paring is not correct, EF-Tu (or EF1) is not released, peptide bond can’t form
* **if base-pairing is correct** GTP is hydrolyzed and EF-Tu/EF1 is released. tRNA is then in the right position, there is a slight delay for one last check before p.p bond formation
\
* ribose and 3 phosphates
* It checks base-paring. if base paring is not correct, EF-Tu (or EF1) is not released, peptide bond can’t form
* **if base-pairing is correct** GTP is hydrolyzed and EF-Tu/EF1 is released. tRNA is then in the right position, there is a slight delay for one last check before p.p bond formation
\
31
New cards
EF-G/EF2
helps the ribosome to move the mRNA forward one codon and helps speed up elongation of the polypeptide chain
* basically moves the small subunit which “moves” the mRNA
* basically moves the small subunit which “moves” the mRNA
32
New cards
Can ribosomes perofrm protein synthesis without the aid of elongation factors?
Yes, but slower and inefficient
33
New cards
Role of elongation factors (2)
improve speed and efficiency
error checking function
error checking function
34
New cards
How are elongation factors mediated?
GTP hydrolysis (breaking it apart to form GDP and Pi) and release of EF-Tu & EF-G
35
New cards
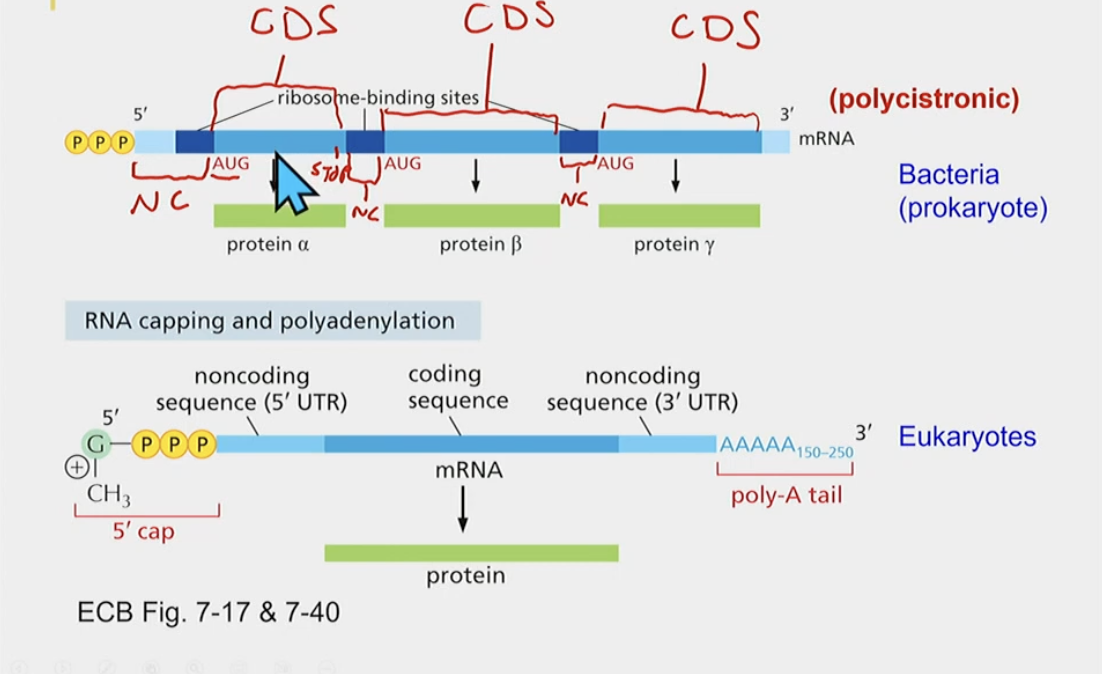
mRNA structure bacteria vs. Eukaryotes
Bacteria:
* ribosome-binding sites and non-coding sequences
* you can have multiple proteins from one mRNA, due to having multiple coding sequences (polycistronic)
* coding sequences, meaning multiple starts and stops
Eukaryotes:
* Monocystronic (codes for one protein) doesn’t have multiple ribosomal binding sites?
* One singular coding sequence creates one protein
* also has 2 protein sequences
* **has a cap and poly-A tail**
* ribosome-binding sites and non-coding sequences
* you can have multiple proteins from one mRNA, due to having multiple coding sequences (polycistronic)
* coding sequences, meaning multiple starts and stops
Eukaryotes:
* Monocystronic (codes for one protein) doesn’t have multiple ribosomal binding sites?
* One singular coding sequence creates one protein
* also has 2 protein sequences
* **has a cap and poly-A tail**
36
New cards
PROKARYOTES: Order of initiation of translation (4 steps)
1. Shine-Dalgarno sequences on mRNA base pair with rRNA in small ribosomal subunits
2. Positioning of small subunits to initiating AUG codons on mRNA also requires Initiation Factors (IFs)
3. fMethionine aminoacyl tRNA binds to initiator codon (AUG)
4. Large ribosomal subunit binds
37
New cards
Shine-Dalgarno Sequences
sequences that allow for binding to rRNA in small ribosomal subunits
38
New cards
EUKARYOTES: Initiation of translation steps
eIF4E (Eukaryotic initiation factor) binds to 5’ cap
eIF4G binds to eIF4E and poly-A binding proteins which is binded to poly-A tail
* Eukaryotic mRNA MUST have a loop for extra quality control check
1. Small ribosomal subunit with translation IFs and INTIATOR TRNA ALREADY BOUND without RNA
* Finds AUG met start codon
* Then large ribosomal subunit comes in
\
eIF4G binds to eIF4E and poly-A binding proteins which is binded to poly-A tail
* Eukaryotic mRNA MUST have a loop for extra quality control check
1. Small ribosomal subunit with translation IFs and INTIATOR TRNA ALREADY BOUND without RNA
* Finds AUG met start codon
* Then large ribosomal subunit comes in
\
39
New cards
Termination of translation
* Specific protein translation release factor recognizes stop codon, mimics tRNA structure to fit into A-site (both eukaryotes and prokaryotes)
40
New cards
Polyribosomes (eukaryotes or prokaryotes?)
**Both euk. and prok.**
A bunch of ribosomes on an mRNA
Relatively slow protein synthesis
one ribosome every \~80 nucleotides
the spiral shape
A bunch of ribosomes on an mRNA
Relatively slow protein synthesis
one ribosome every \~80 nucleotides
the spiral shape
41
New cards
Protein folding (chaperone proteins)
Proteins that help other proteins fold
exaples of molecular chaperones:
* Hsp60
* Hsp70
Hsp stands for heat shock protein
* when heat shock causes protein to denature, these proteins help the protein fold back up
exaples of molecular chaperones:
* Hsp60
* Hsp70
Hsp stands for heat shock protein
* when heat shock causes protein to denature, these proteins help the protein fold back up
42
New cards
Post-translational modifications (list them and why they may be required)
Many proteins require post-translation modifications such as:
* phosphorylation
* glycosylation
Covalent modifications may be required to:
* make protein active
* recruit protein to correct membrane or organelle
\
* phosphorylation
* glycosylation
Covalent modifications may be required to:
* make protein active
* recruit protein to correct membrane or organelle
\
43
New cards
Control of protein degradation
Some proteins are short-lived, while others may last for months or years.
**Proteins targeted for degradation** have a small protein called **ubiquitin** covalently attached to them, (creating a polyubiquitin chain) which **directs them to the proteasome** where they are **degraded by proteases**, and the **amino acids recycled into new proteins in the cell, ubiquitin is recycled as well, not destroyed**
**Proteins targeted for degradation** have a small protein called **ubiquitin** covalently attached to them, (creating a polyubiquitin chain) which **directs them to the proteasome** where they are **degraded by proteases**, and the **amino acids recycled into new proteins in the cell, ubiquitin is recycled as well, not destroyed**
44
New cards
Antibiotics (what they are) and modes of action (3 points total)
1. Antibiotics: act on **prokaryotes**
2. **Not all,** but many inhibit **translation**
1. acting on **ribosome**
3. **Drugs**
\
Things that act on both bacteria and eukaryotes or just eukaryotes are basically just poisons for us
45
New cards
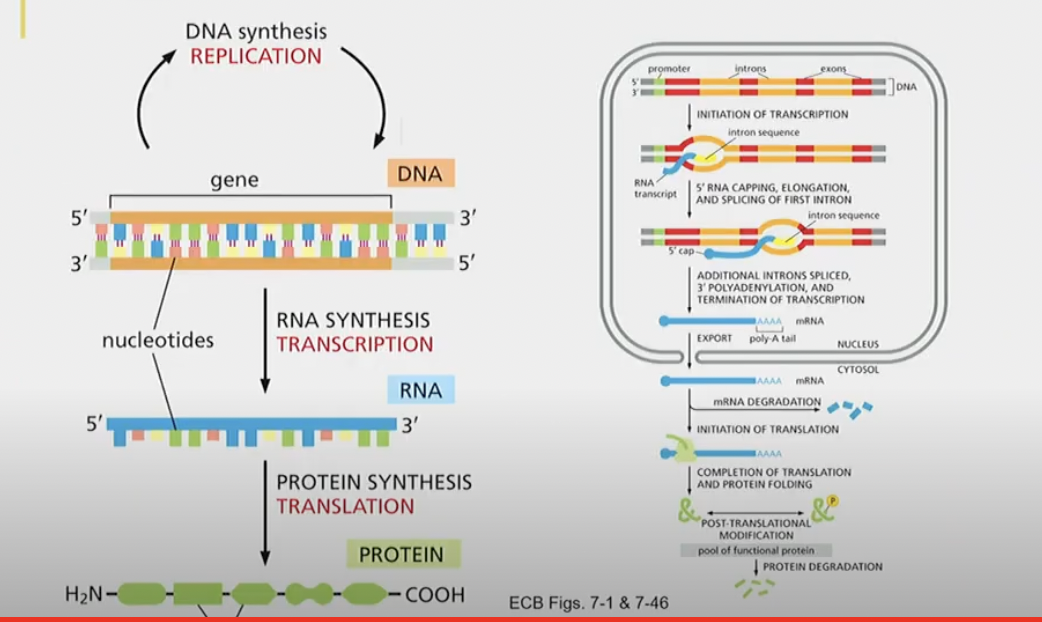
Describe information flow. Transcription to translation, the whole shebang.