Secretion systems in bacteria
1/23
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
24 Terms
What is bacterial secretion and why is it important?
The active transport of molecules from the cytoplasm to the extracellular environment by specific protein systems
It’s essential for virulence (e.g., toxin secretion), nutrient acquisition, cell communication, and biofilm formation.
In biotechnology, secretion allows production and release of recombinant proteins.
Secretion plays a key role in symbiosis as well — for example, rhizobia secrete nodulation factors to communicate with plants.
Differentiate between secretion, export, and translocation.
Secretion: Active transport from the cell interior to exterior.
Export: Active transport from cytoplasm to i.e. the periplasm (in GN bacteria).
Translocation: Transport across a single lipid bilayer.
Note: “Secretion” in Gram-positive bacteria usually equals “export” in Gram-negative bacteria because they have only one membrane to cross.
What are the secretome and exoproteome?
Secretome: All proteins (i.e translocation system components) secreted by a cell and the secretion machinery.
Exoproteome: The subset of those proteins found extracellularly.
Advanced detail: The secretome is a major part of bacterial adaptation — e.g., pathogens have a larger, more diverse secretome than commensals.
What are the major structural barriers to secretion in bacteria?
Monoderm bacteria (Gram-positive): One membrane + thick peptidoglycan wall.
Diderm bacteria (Gram-negative): Inner and outer membranes with a periplasmic space.
👉 Extra: Mycobacteria are atypical diderms with a waxy, mycolic acid-rich outer layer (“mycomembrane”), creating unique secretion challenges.
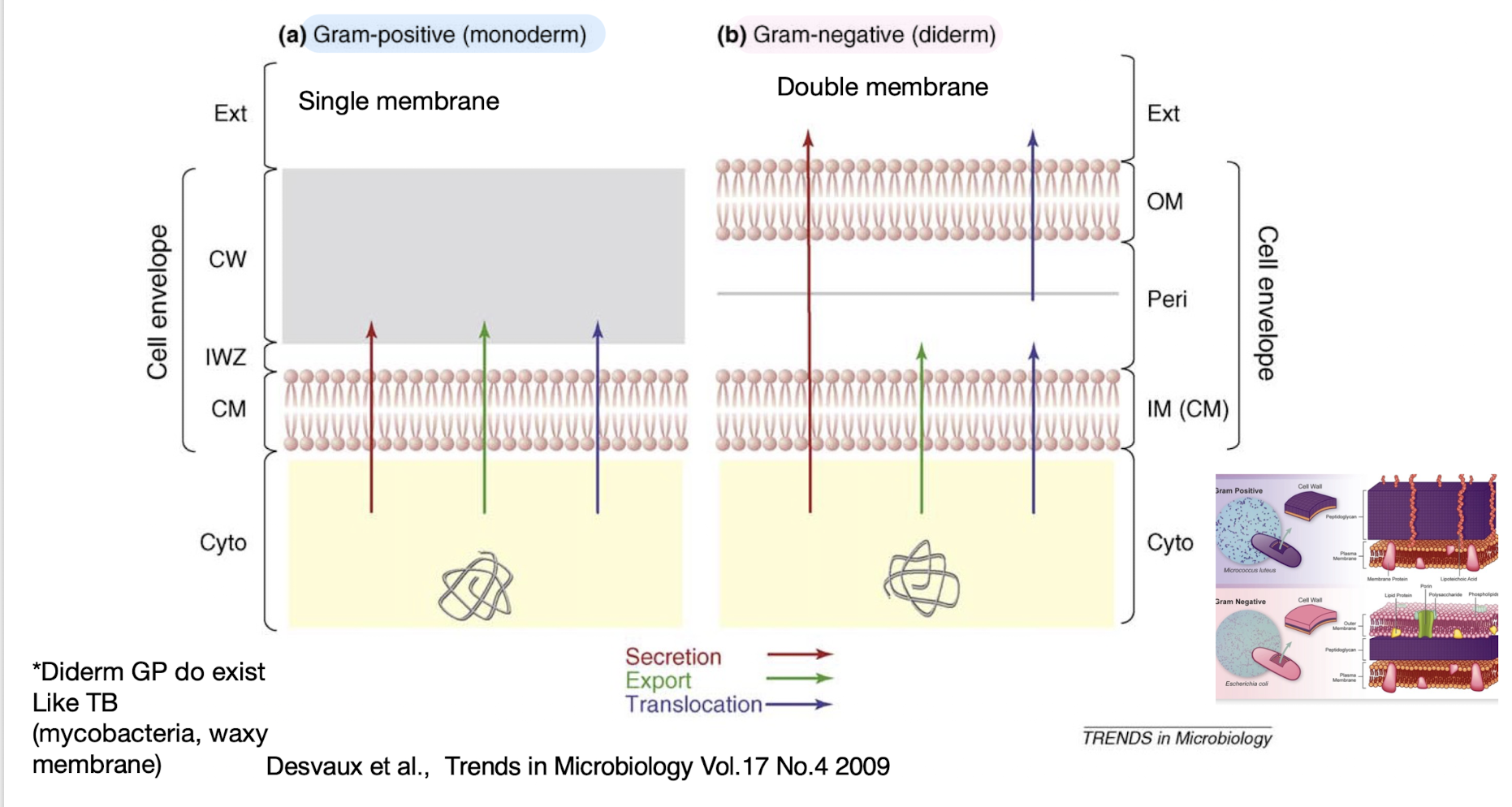
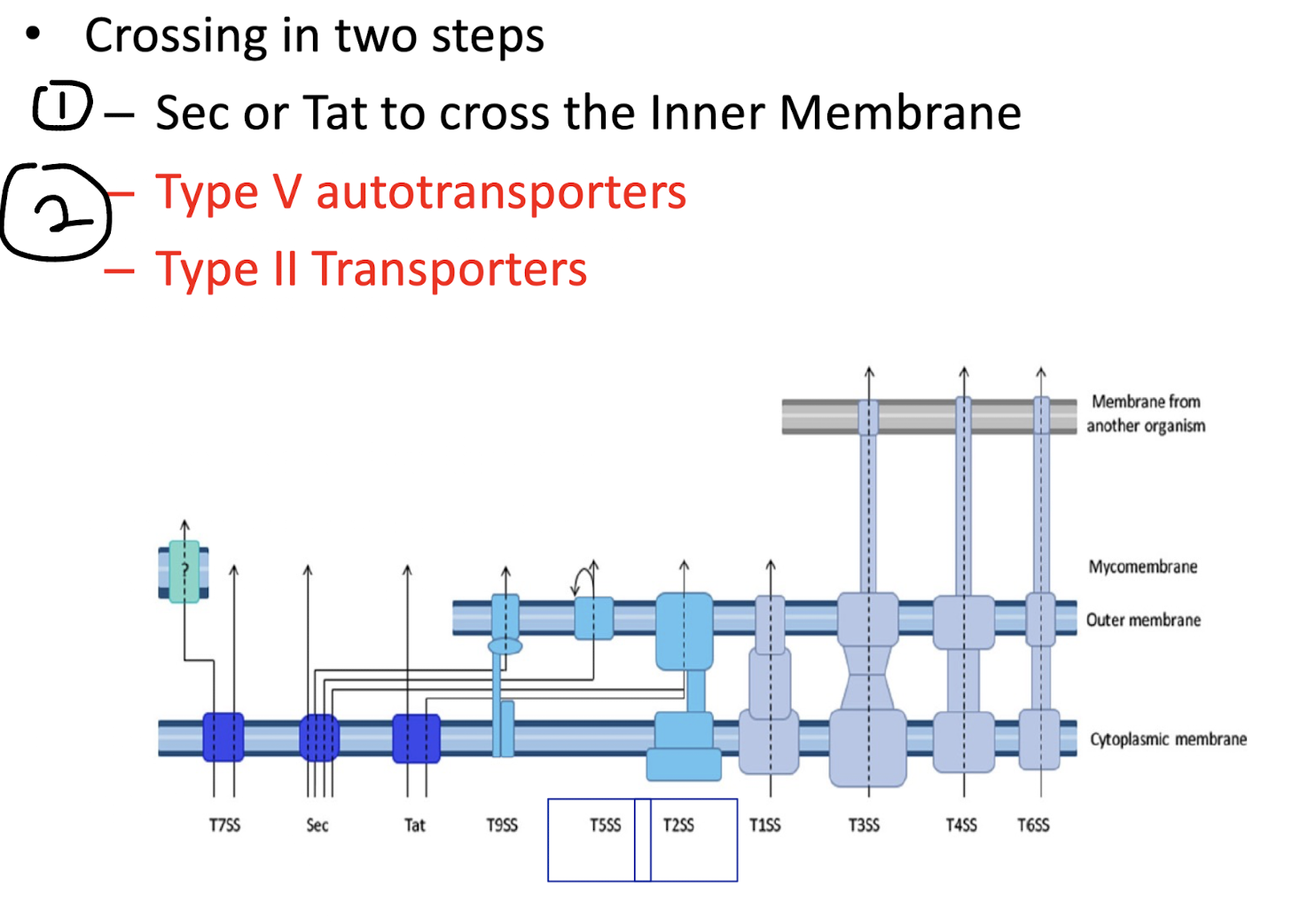
Monoderm bacteria primarily use the ______
Sec or Tat pathway and the T7SS
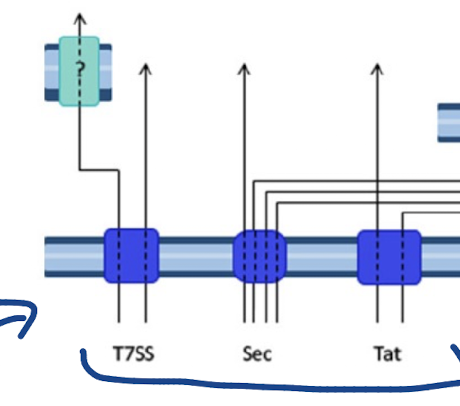
Diderm non-LPS bacteria primarily use the ______ while diderm-LPS use ______
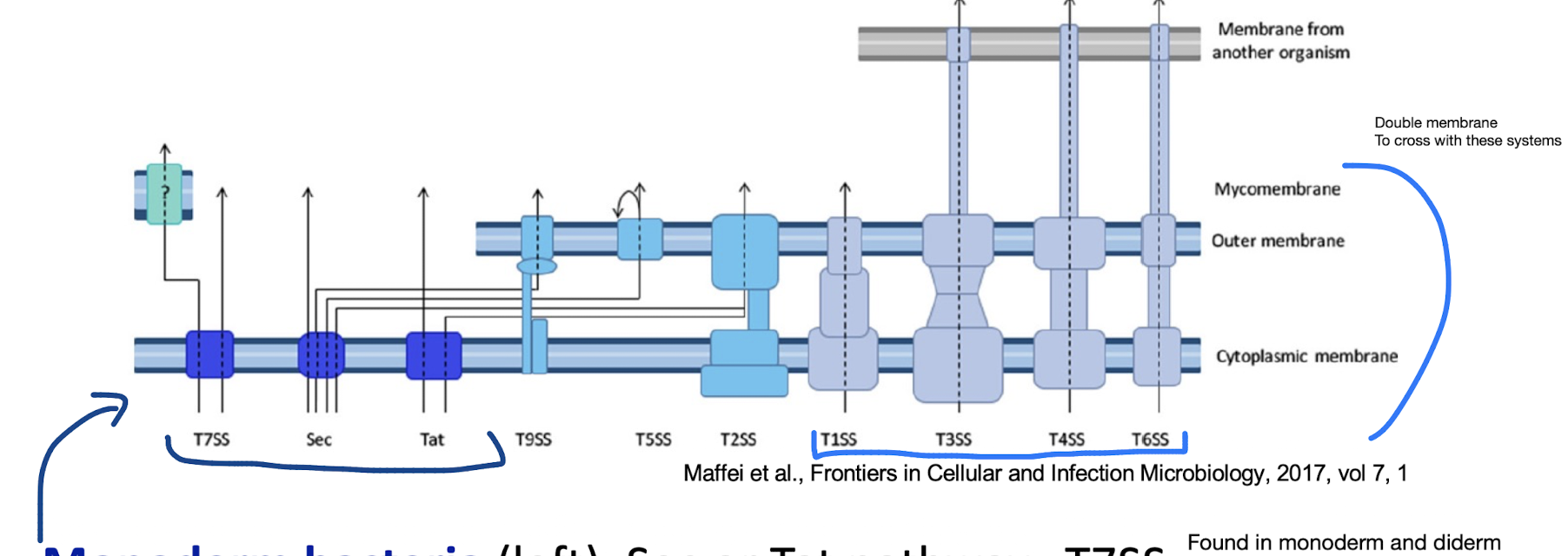
Diderm-non-LPS bacteria such as Mycobacteria (far-left), T7SS
Diderm-LPS bacteria (centre and right), T1SS, T3SS, T4SS, or T6SS.
Other secreted proteins are first exported to the periplasm via the Sec system (T2SS, T5SS, or T9SS) or the Tat system (for T2SS only).
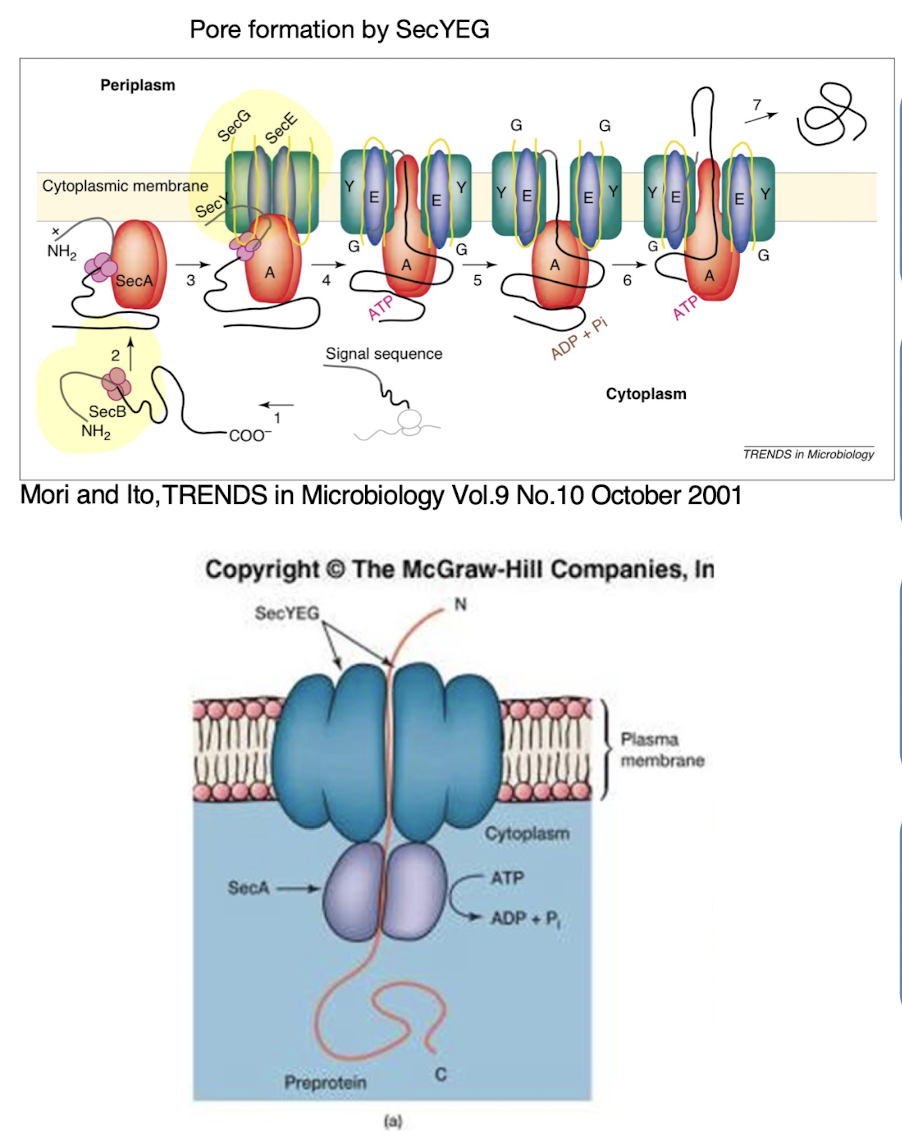
Describe the Sec secretion system and its energy requirements.
The Sec system exports unfolded proteins across the cytoplasmic membrane using ATP hydrolysis (via SecA) and sometimes PMF
SecYEG: forms the pore where the protein travels through
SecB: recognises signal peptide and guides protein to SecYEG.
SecA: ATPase / molecular pump
Signal peptide is cleaved post-translocation and then undergoes folding
- Found in GP bacteria where only one membrane needs to be passed as it passes the sec translocase system
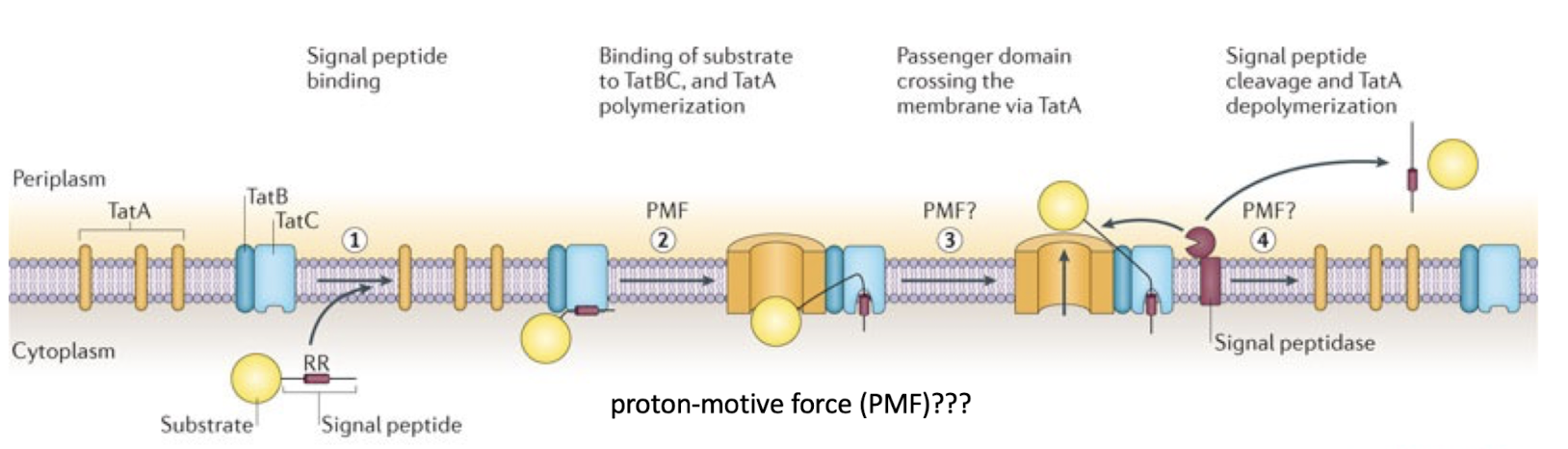
How does the Tat system differ from the Sec pathway?
Translocates folded proteins across the membrane.
TatBC recognises a twin-arginine residue motif (RR) in the signal peptide.
Driven by proton motive force, not ATP.
Not as wide spread as the sec system
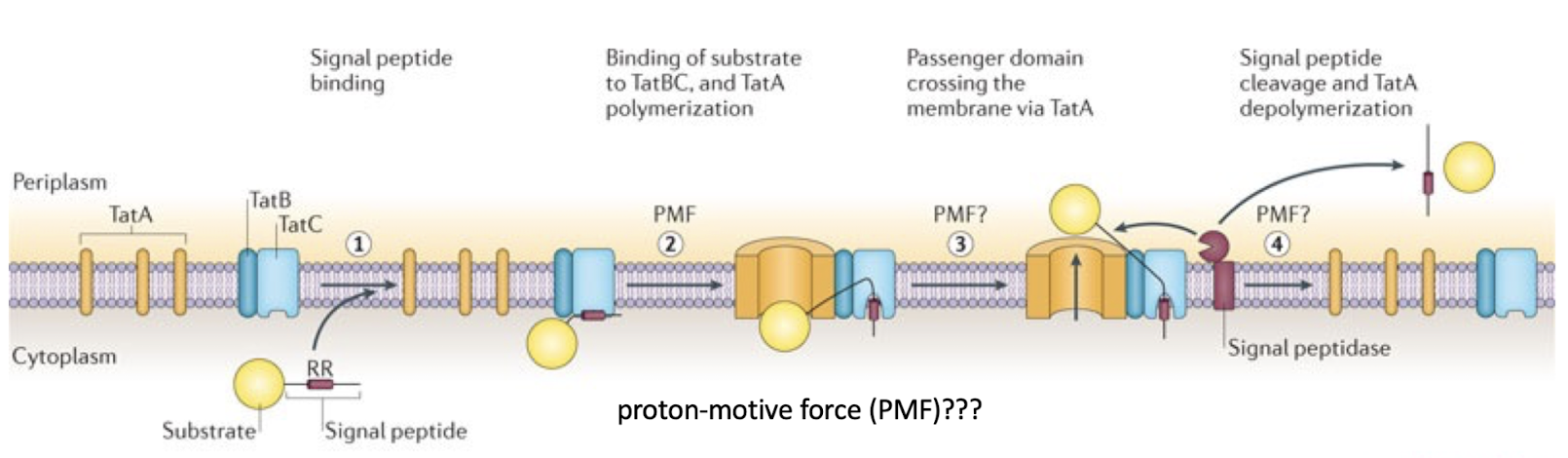
Why does TatA appear to weaken the membrane during transport?
TatA oligomerizes to form a dynamic pore, expanding to accommodate folded proteins. This transient “loosening” facilitates passage of large substrates.
The Tat pore is not permanently open — it disassembles after each transport event, minimising proton leakage.
Proton flux through the pore formed may aid in the transport of the protein
Why can Sec and Tat systems alone only partially achieve secretion in Gram-negative bacteria?
Because they only translocate proteins into the periplasm, not across the outer membrane.
Additional systems (Type II, V, IX (nine), etc.) are required to complete the process
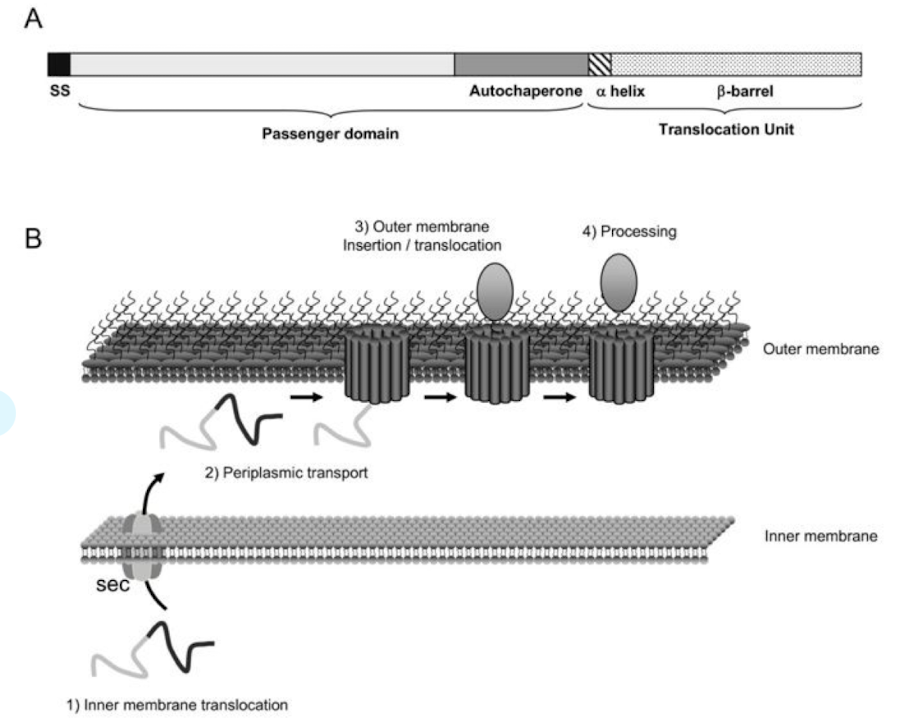
How do T5SS autotransporters function?
Classical autotransporters
– single polypeptides that contain a signal peptide that directs transport over IM via Sec
– N-terminal extracellular domain (‘passenger domain’)
– passenger domain can function as hydrolases, cytotoxins, or adhesins, or have other activities associated with virulence
– C-terminal domain that resides in the outer membrane (OM) with the help of the Bam (β-barrel assembly machinery) complex.
This domain folds into a β-barrel structure that forms a channel or pore through which the N-terminal part of the protein (the passenger domain) is pushed to the cell surface.
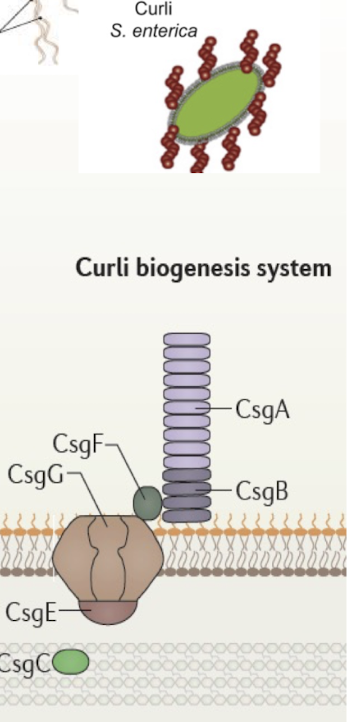
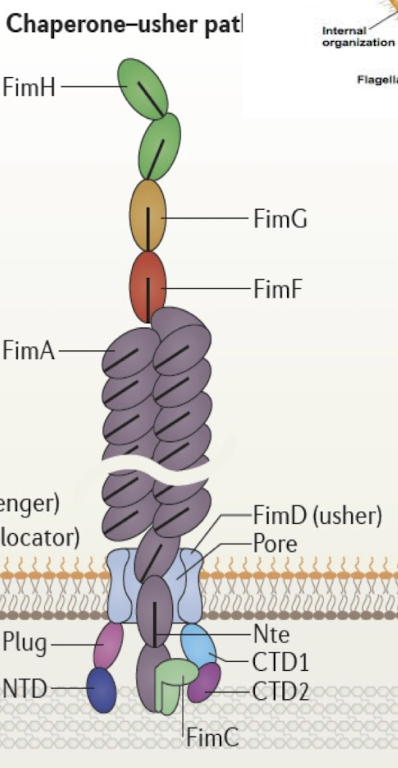
What is the Chaperone-Usher system and what is it similar to?
Assembles and secretes adhesive surface appendages (pili/ fimbriae)
Chaperone (FimC) stabilises pilin subunits in the periplasm.
Usher (FimD) forms OM channel for fiber assembly.
Tip formation (FimHGF) followed by 1000s of subunits (FimA)
This pathway is crucial for host colonisation and biofilm formation — a target for anti-adhesion therapies.
Similar to the Curli biosynthesis in S. enterica with CsgE forming the pore
How does the Type II secretion system operate?
Inner membrane complex forming a platform beneath GspD,spans both IM + OM
Receives proteins already exported to periplasm via Sec/Tat.
Uses GspE ATPase for energy.
GspD forms large OM pore allowing folded proteins through
GspG pseudopilus acts as a piston pushing proteins out.
May interact with cytoskeleton
Advanced point: Functionally analogous to type IV pili — in fact, evolutionarily derived from them

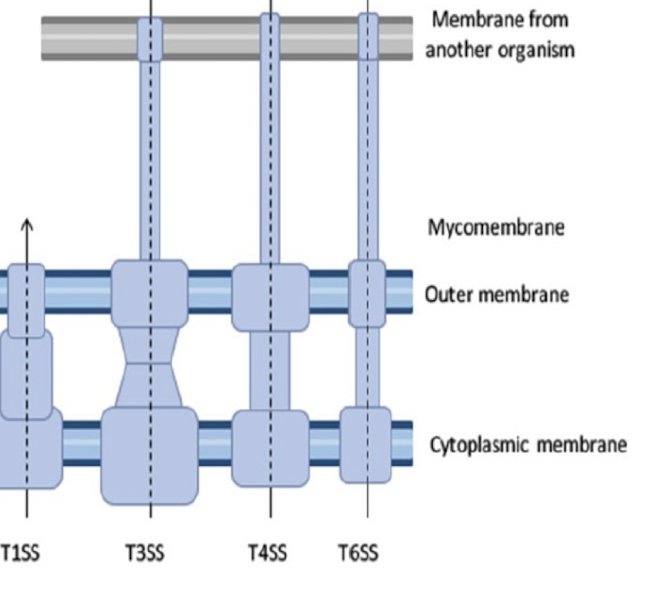
Which secretion systems can transport directly from the cytoplasm to the extracellular space in a single step?
Type I, III, IV, and VI.
These systems form continuous channels across both membranes, bypassing the periplasm.
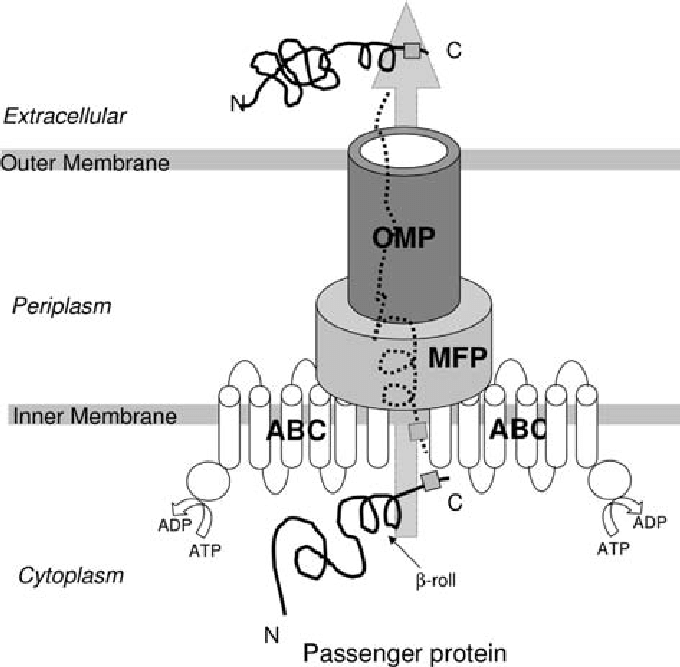
What are the key components and energy drivers of T1SS?
ABC transporter in IM (ATP-binding) , transports proteins in a single step in GN bacteria
Membrane fusion protein (MFP) connecting IM and OM.
OM protein of TolC family
Recognises unfolded proteins via C-terminal uncleaved signal sequence
Driven by ATP and proton motive force.
Example: Hemolysin secretion in E. coli.
How does the Type III secretion system function and why is it important?
Complex structure acting as a molecular syringe injecting effector proteins directly into host cells - hence also known as “the injectisome”
Requires chaperones to transport unfolded effector proteins to the needle
Evolutionarily related to flagella.
Pathogens like EHEC, EPEC, Salmonella, Shigella, and Yersinia use T3SS to manipulate host actin and signaling pathways for invasion.
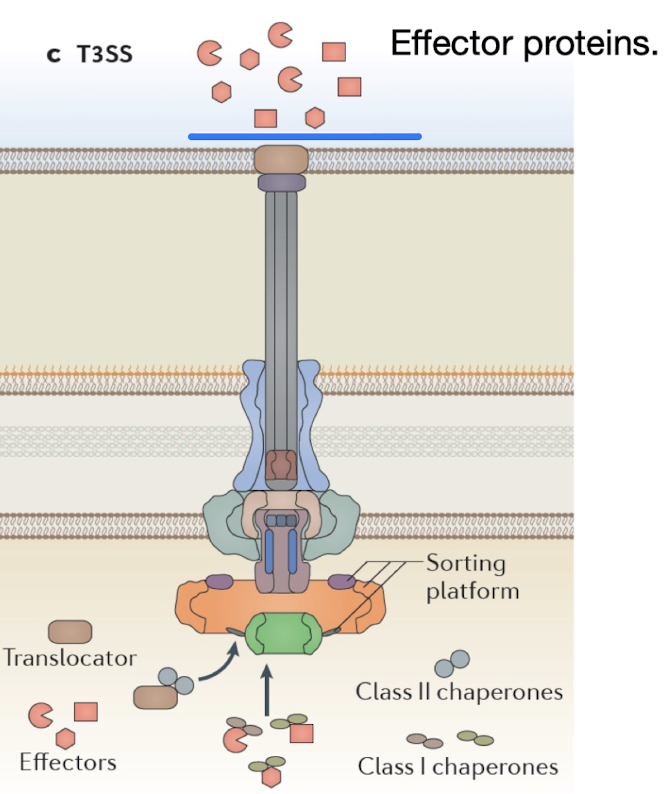
How do pathogens like pathogenic E. coli pathotypes manipulate host actin in order to invade the cell?
Pathogenic E. coli pathotypes utilize the Type III secretion system (T3SS) to inject effector proteins e.g. Tir into host cells, which manipulate the host's actin cytoskeleton by promoting actin polymerisation and forming protrusions that facilitate bacterial entry.
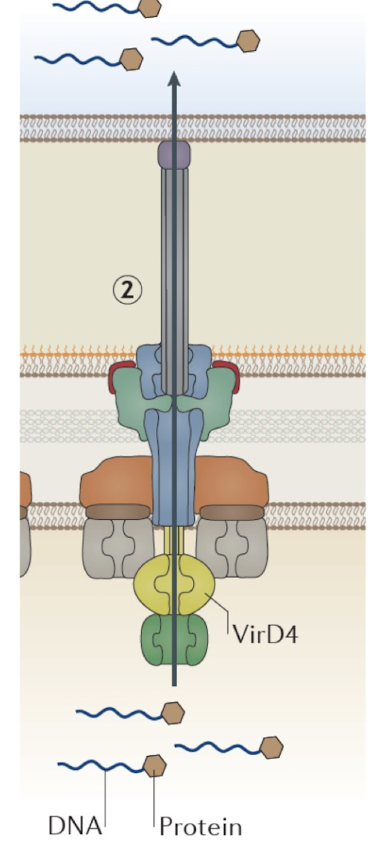
What is the T4SS known for?
Can transfer DNA or proteins (e.g., conjugation or effector delivery).
Acts similarly to T3SS
Found in Agrobacterium tumefaciens (T-DNA transfer to plants).
Extra: T4SS is central to horizontal gene transfer and antibiotic resistance dissemination.
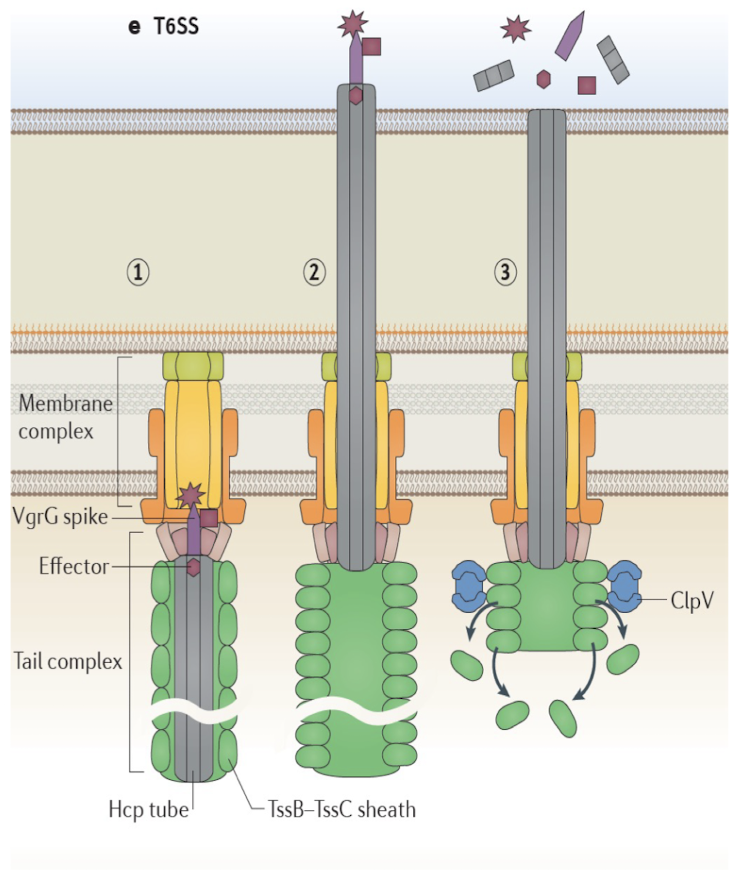
Describe the structure and function of T6SS.
Membrane complex similar to Type IV SS
Tail complex similar to bacteriophage tail.
Signal event from effector protein leads to movement of the Hcp tube and consequent injection of protein
Often used in bacterial warfare (interbacterial competition).
👉 Extra: Vibrio cholerae and Pseudomonas aeruginosa use T6SS for both virulence and niche dominance.
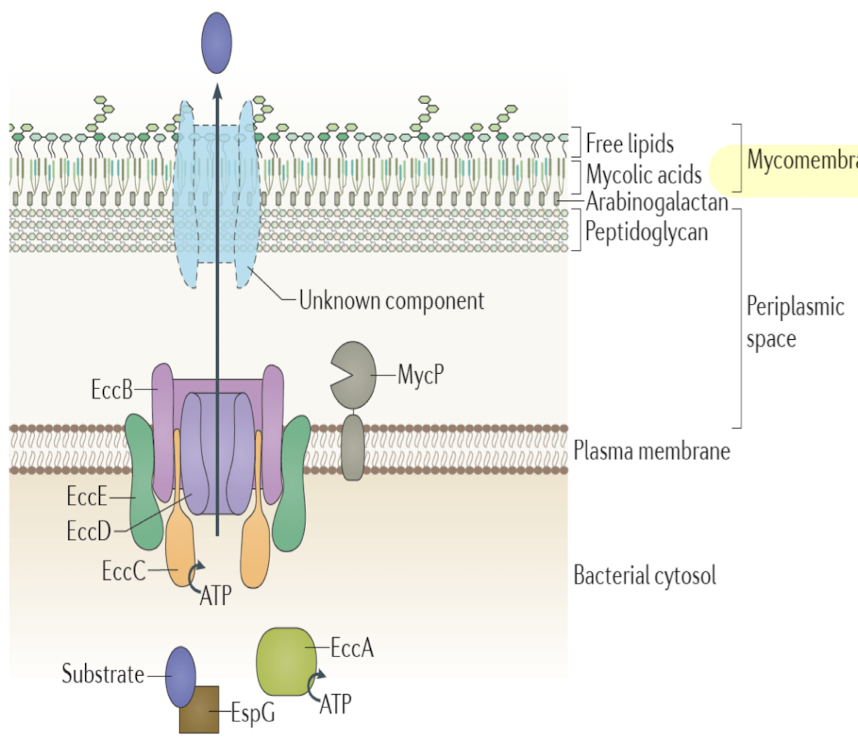
What makes T7SS (Type VII) unique and where is it found?
Found in mycobacteria and Gram-positive diderms.
Transports virulence factors across the mycomembrane instead of OM
Mycomembrane consists of waxy mycolic acids and carbohydrates, confer antibiotic resistance
One core IM channel defined, direct mechanism of how substrates transverse mycomembrane is not clear
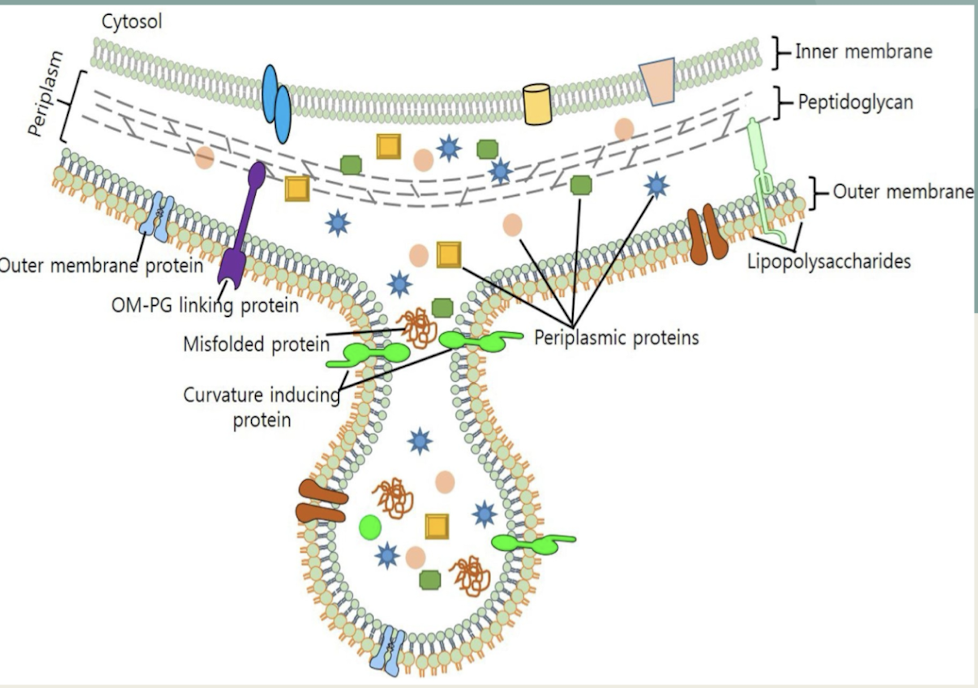
What are OMVs and how do they contribute to bacterial physiology?
Spherical vesicles blebbed from OM in Gram-negative bacteria.
Contain periplasmic proteins, toxins, signaling molecules.
Facilitate horizontal gene transfer, immune evasion, and host manipulation.
How are secretion systems evolutionarily related to other bacterial structures?
T3SS evolved from flagellar basal body.
T4SS from conjugation pili systems.
T6SS from phage contractile tails.
Advanced: This evolutionary modularity highlights how bacteria repurpose molecular machinery for new ecological and pathogenic roles.
How do secretion systems contribute to bacterial virulence and survival?
Direct toxin delivery into hosts (T3SS, T4SS, T6SS)
Adhesion (autotransporters, pili)
Immune evasion (OMVs)
Interbacterial killing (T6SS)
👉 Extra: Secretion systems are often encoded on pathogenicity islands, indicating horizontal acquisition during adaptation to pathogenic lifestyles.
What pathogenicity island is common in pathogenic E.coli?
The Locus of Enterocyte Effacement (LEE) is common in pathogenic E. coli and is crucial for its virulence.