lf206 lecture 4 - eukaryotic genomes, histones
1/27
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
28 Terms
approx. 40% of our genome is derived from ______________.
what are they?
retrotransposons
can copy and paste themselves using RNA intermediate
why do we have multiple haemoglobin genes?
allows for gradual change from foetal to adult haemoglobins - requires temporal transcriptional regulation
protein-coding genes have relatives of which they share common _______.
some genes exist in families and _____-families
some are pseudogenes - what are these?
within a genome, families can be dispersed or c_______. maintenance of clusters implies _________
ancestry
super-families
non-functional copies of a gene - they look like the gene but are not functional.
functional co-ordination/regulation
expressed genes are found in what formation?
euchromatin
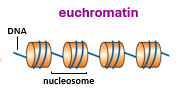
genes not expressed are usually found in what conformation?
facultative heterochromatin
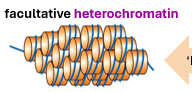
difference between facultative and constitutive heterochromatin
facultative can switch into euchromatin - so is not always packed
constitutive is permanently not expressed - these genes are silent.
three specific structures of constitutive heterochromatin
origins of replication
telomeres
centromeres
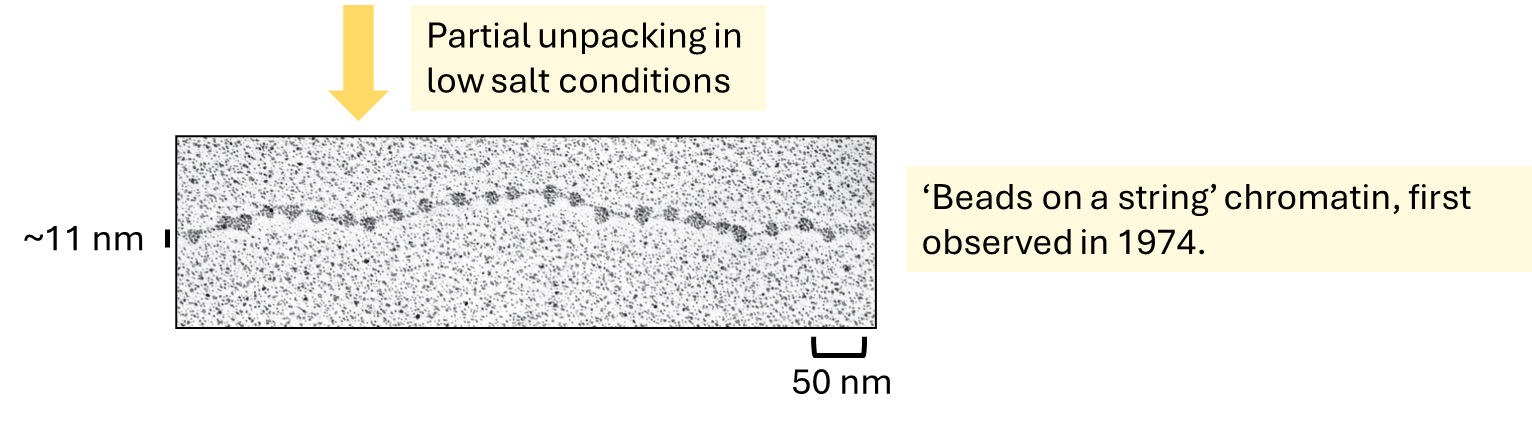
what is this?
euchromatin
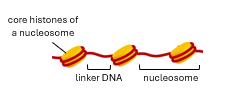
what is a nucleosome?
147bp dsDNA wrapped around a octameric histone core (of 8 histone proteins)
how many and what histone subunits per nucleosome
8 subunits
2x H2A
2x H2B
2x H3
2x H4
how to dissect nucleosome core particle to release histones
digest linker with DNAse
high salt dissociation to remove octameric histone core and 147bp dsDNA
rebuilding euchromatin/nucleosome
express each histone separetly in E.coli
purify each histone
combine in different combination to explore interactions
H3 and H4 interact to form a dimer, which binds to a tetramer, which can bind DNA
palindromic dsDNA sequence of 146bp on a recombinant plasmid maintained in E.coli
H3-H4 tetramer:DNA complex binds 2 separate H2A:H2B dimers
reconstituting nucleosomes in vitro which pack evenly during crystallisation = high-resolution structure
nucleosomes are not static. they are ___________ to make DNA more/less accessible to ______ proteins, general transcription ______, mediator ______ and RNA _________ II
thus histone modification ______ transcription
remodelled (removed/re-positioned/replaced/covalently modified)
activator
factors
proteins
polymerase
regulate
histone tails can be modified.
specific ______ (K) residues can be modified by _________ (Ac).
this is a code to ‘_____ the chromatin structure’
lysine
acetylation
relax
how can a gene be silenced?
some _____ (K) and arginine (___) residues can be modified by _________ (Me) on the histone ____/tail
this can relax or compact chromatin depending on
which residue is methylated
the ________ of methylation (mono-, di-, tri-)
lysine
R
methylation
code
degree
phosphorylation (P) of some _____ (S) and _______ (T) residues promotes ________
____________ (Ub) of some _______ (K) on H2A are __________ sites for DNA ______
serine
threonine
transcription
ubiquitylation
arginine
recruitment
repair
define epigenetics
heritable changes in gene expression that are not mediated at the DNA sequence levels
(e.g. DNA modifications, histone modifications)
how can histone modifications be maintained during transcription
epigenetic marks are preserved
RNA Pol II transcribes naked DNA quickly but transcribes ‘beads on a string’ slowly with pauses
RNA Pol II stalls at nucleosome A
RNA Pol II unwinds about half of the DNA from nucleosome A
transcribed DNA is looped and starts to wind around nucleosome A whilst unwinding continues
identifying origins of replication in yeast
ARS region?
autonomously replicating sequences
DNA sequences that can replicate independently and are functional origins of replication
origin of replication?
site on DNA where replication begins
cell cycle phases
INTERPHASE
G1
S
G2
MITOTIC PHASE
mitosis
cytokinesis
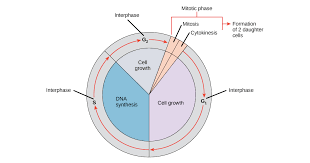
G1
an origin of replication has a pre-bound complex of ___ proteins (_______). they remain ______ throughout the cycle
Cdc6 and Cdt1 associate with ___
___ helicases are recruited to form the _______ ______ (pre-RC)
in G1 pre-RC is formed but not _______
ORC (ORC 1-6)
associated
ORC
Mcm
prereplicative complexes
activated
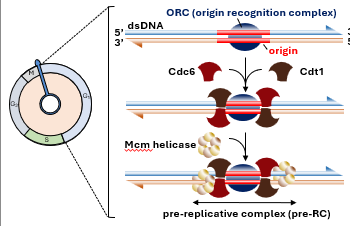
S phase
Cdk2 (_____ ________ ____ 2) phosphorylates ____
proteasomal __________ of phosphorylated Cdc6
phosphorylation of ___ = RC is activated but new ones cannot form (no Cdc6)
Mcm _____ open the DNA allowing for recruitment of primase clamp _____ and DNA _______.
cyclin dependent kinase 2
Cdc6
degradation
helicases
loader
polymerases
what happens to histones during replication
replication machinery displaces H2A-H2B dimers.
unclear whether H3-H4 tetramers are displaced or reused?
but both new strands inherit them
large increase in expression of histones in S phase
new subunits acetylated to promote open structure
NAP-1 (nucleosome assembly protein 1) chaperones H2A-H2B dimers
CAF-1 (chromatin assembly factor) chaperones H3-H4 tetramers
assembles nucleosomes in an open chromatin structure
fresh acetylations are removed, ‘reader-writer complex’ copies epigenetic information from neighbouring nucleosomes.
thus daughter cells maintaining parental cell histone modifications
cytosines in DNA can be methylated.
methylated DNA means __ ___ _____
methylated histones can recruit DNA _____ that methylate _____ residues
methylated DNA can recruit histone _______ = ______ by condensing chromatin
histone code and DNA methylation work together
do not express
methylases
cytosine
methylases
silencing
what is SINE
short interspersed nuclear elements
80-500bp long
a type of retrotransposon
how can SINEs regulate gene expression (non-epigenetic and epigenetic)
non-epigenetic: act as enhancers or alternative promoters to recruit transcription factors & promote expression
epigenetic: SINEs are GC-rich so hotspot for DNA methylation, silence nearby genes by stimulating chromatin condensation