GENET 270: Lec 3 - Bacterial chromosomes pt. 2
1/78
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
79 Terms
Is the bacterial chromosome more or less structured than eukaryotic chromosome?
Less structured
True or false: Many rounds of replication can occur at the same time?
True
multiple copies of genes present when new rounds of replication initiate before other rounds finish
****What does the simultaneous rounds replication result in?
POLYPLOIDY until cell division catches up to DNA replication
Do bacterial chromosomes use telomeres?
No
not linear therefore don’t need buffer of degradation
***Where does replication initiate for bacteria?
Specific site = OriC
How many replication forks are there for bacterial replication?
2
bidirectional
***What is the typical size range of oriC?
Less than 260 bp of DNA
*****What does oriC contain?
Binding sites for proteins required to initiate replication
Many DnaA Binding sites
IHF binding sites
Fis binding sites
****What does DnaA do?
DNA unwinding element/ opens Helix
***What do IHF + Fis each do?
Regulate DNA replication
Integration host factor = BENDS dna
Fis= prevent premature DnaA binding to for multimer
*****Where are the 2 places DnaA can bind on the oriC?
High affinity sites
I, sigma + DUE sites
****When is Dna A bound to each site?
DnaA = ALWAYS bound to High affinity sites
Only ATP-bound DnaA can bind to I, sigma + DUE sites
*****Why does Replication only occur when the Energy stores of the bacteria are favorable
Only ATP bound DnaA can bind to the I, sigma and DUE sites
When these sites are bound it forms a large MULTIMER
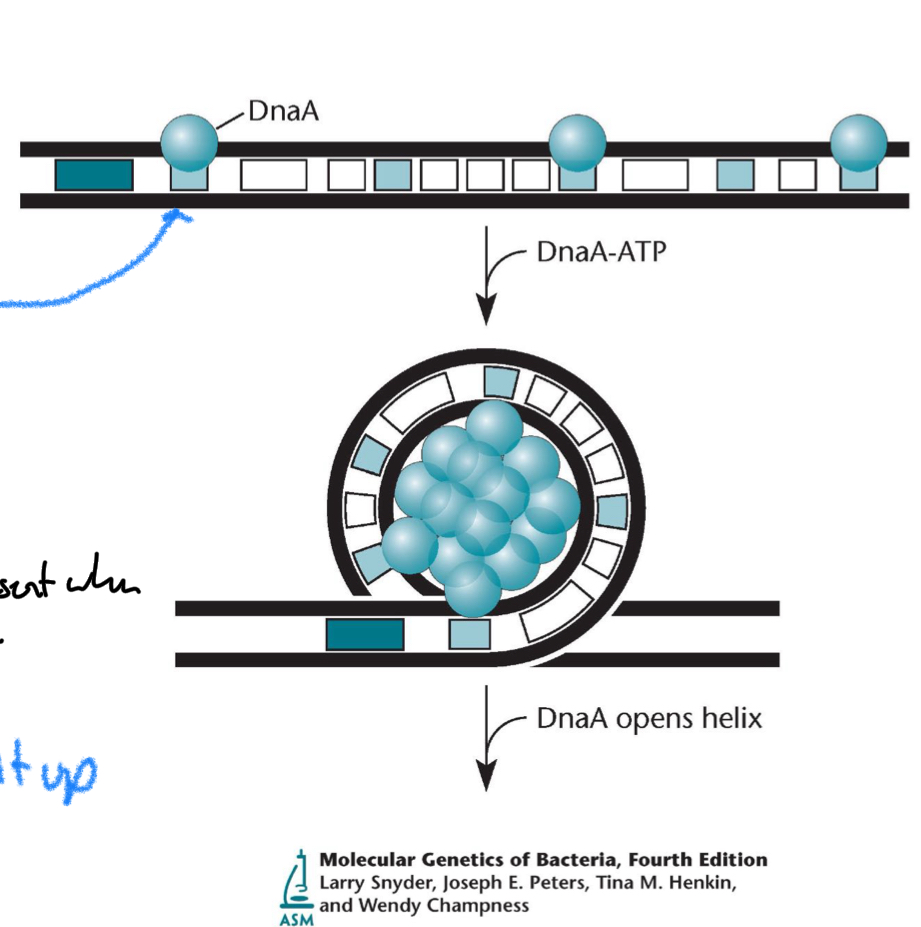
*****List the process by which DnaA opens the helix
DnaA-ATP binds oriC
DnaA forms a multimeric filament
Fis vs. IHF regulation
Fis bound at oriC prevents too-early DnaA assembly → acts like a checkpoint.
IHF sharply bends DNA (~160°), bringing distant DnaA boxes into proximity.
DNA bending + DnaA multimerization
DnaA-ATP filament wraps the oriC DNA, causing torsional strain.
Opening of AT-rich region
The strain unwinds the AT-rich DNA next to the DnaA boxes.
This creates the open complex.
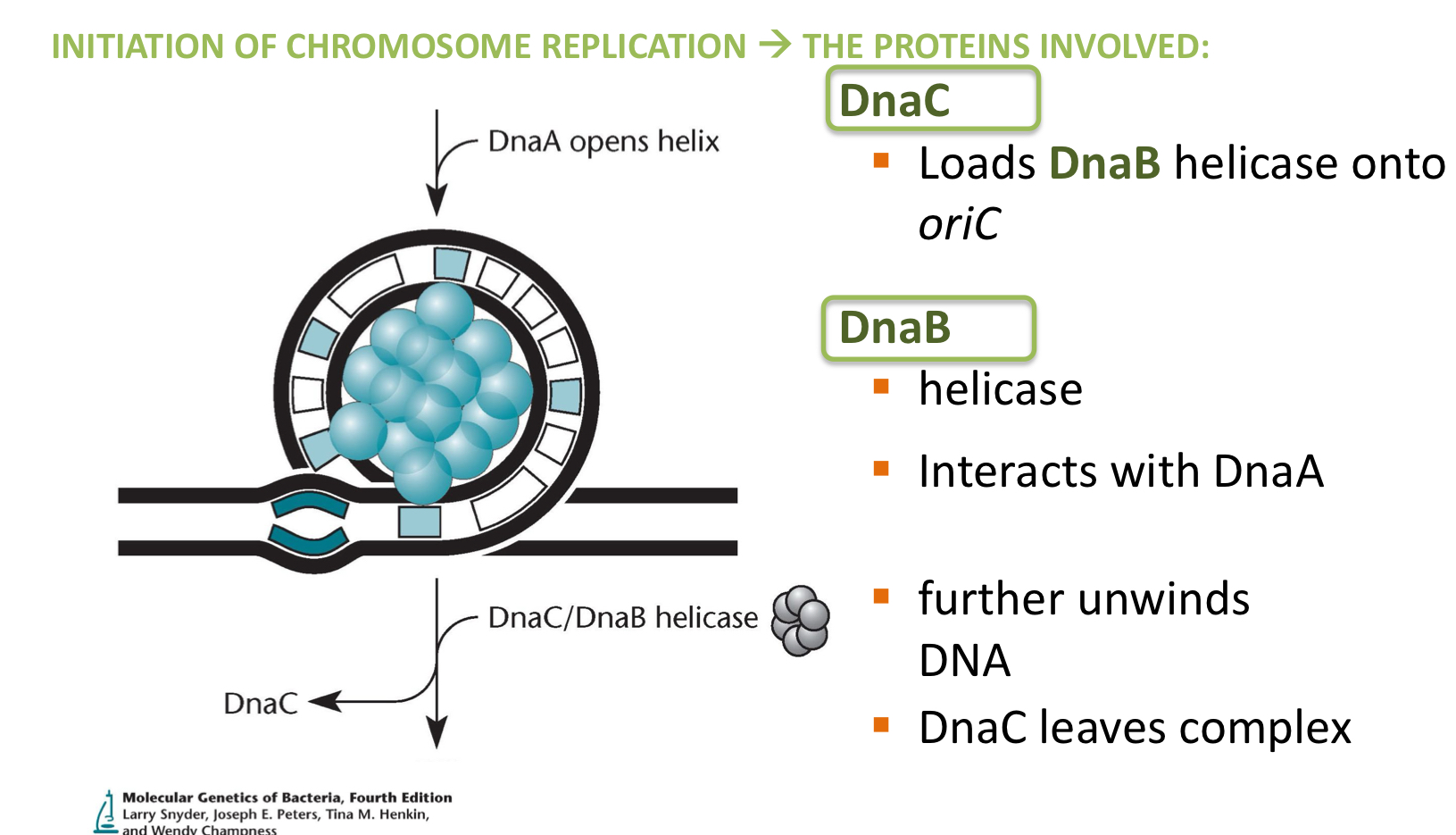
***After DnaA opens up the helix @ oriC, what 2 enzymes come to further open the helix?
DnaC
DnaB
***What does DNA C + B each do during initiation?
C = Loads DNA B onto OriC
B = Helicase, interacts with DNA A + further unwinds the DNA
***What is the Next Enzyme that comes into play with initiation after A B+ C?
DNAG
****What is DNA G + What does it do?
DNA G = PRIMASE
Lays down RNA primers
***What is the PRIMOSOME made of + what does it do?
helicase–primase complex in bacteria (DNA G + B)
Job is to unwind DNA and then synthesize RNA primers so replication can begin
****What is an alternative to DNA G/ Primase when it is not available?
RNAP/ RNA poly
it can lay down the first primer instead of DNA G
****What is the other function of RNAP?
can transcribe through oriC to help open up DNA for DnaA binding
****What all the enzymes involved with Initiation?
DnaA = binds to high affinity sites + I, sigma + DUE sites = forms multimer that creates tension + pulls helix apart (with the help of IHF + Fis)
DnaC = Loads DnaB
Dna B = Helicase
Dna G = Primase (makes RNA primers on DNA)
Primosome = Dna G + B
What are the 2 ways that DNA rep of bacterial chromosomes terminate?
At Ter sites
When replication forks run into each other
***How long are Ter sites?
22 bp long
*****How do Ter sites work?
Act as traps
Replication fork can only pass through in ONE DIRECTION
Like the spikey tire trap speed bumps
******Which Ter sites block what direction of travel? (there are 9 ter sites)
A, D, H + I = Allow Clockwise movement but not counterclockwise (on the right side of the circle pointing clock wise)
B, C, F, G + J = Allow counter clockwise not clockwise (on left side of circle pointing counter clockwise)
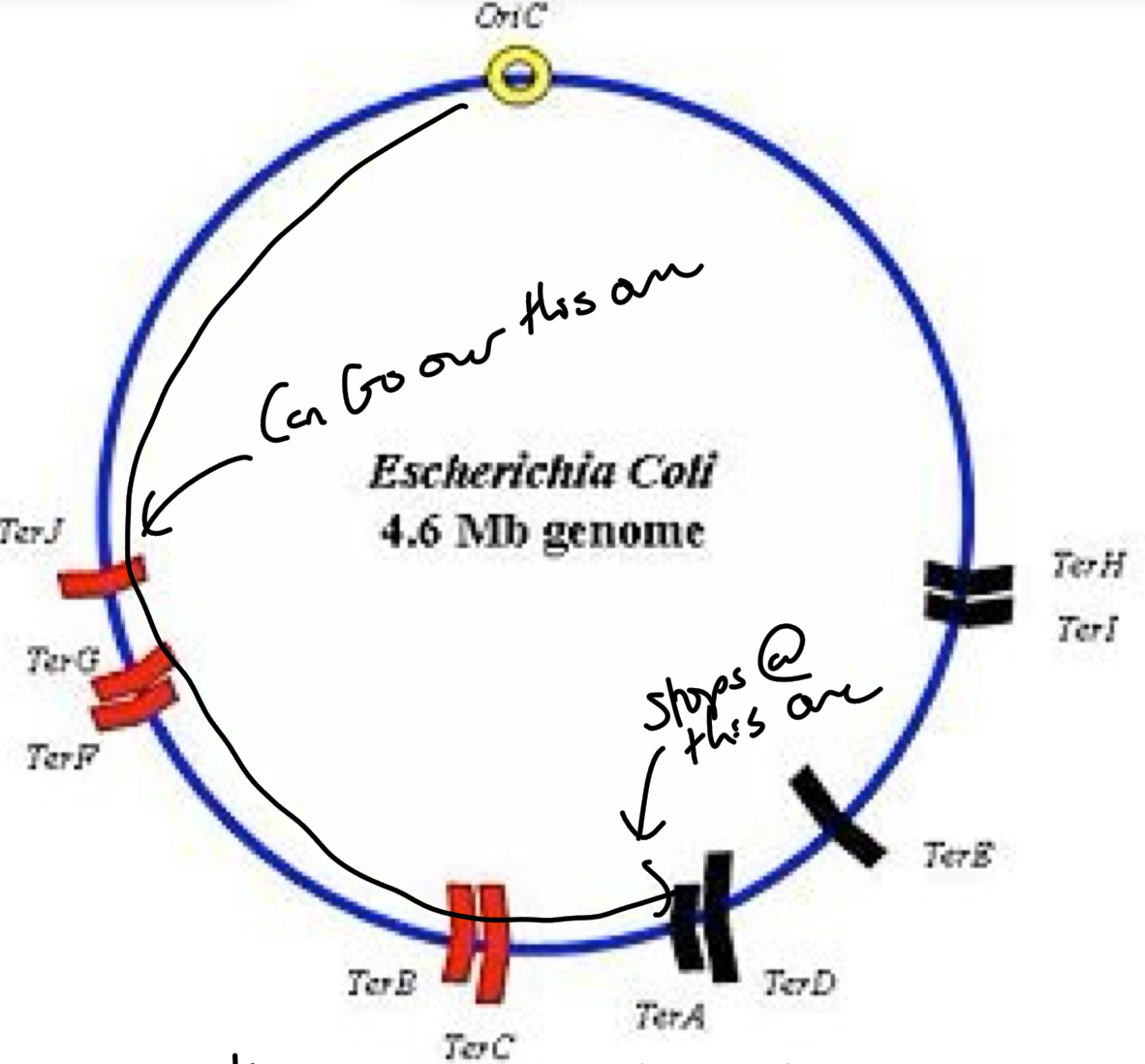
*****What is the purpose of ter sites?
Ensure replication forks meet + terminate at discrete locations
in case of stalled DNA poly, don’t want the the other DNA poly to go into area of replication where head on collision with RNA poly can occur
******In what scenarios are Ter sites used and in what scenarios do the forks run into each other for termination?
Ter sites = Stalling of replication for or the 2 DNA poly not in sync
can cause head on collisions with RNA poly destroying DNA
Run into each other = Replication forks = at same speed
****What must occur before the septum formation + partitioning into daughter cells?
Chromosome segregation
****What 3 things Complicate Chromosome Segregation?
Large size of chromosomes
Possible joining of replicated chromosomes by recombination
Tangling
How was chromosome segregation studied?
Using GFP
****What are Chromosome Dimers?
Double length circular chromosome joined by recombination = PREVENT SEGREGATION
Complicate segregation #2: Possible joining of replicated chromosomes by recombination
***WHAT resolves the Chromosome dimers caused by recombination?
Xer Recombinase at dif sites
****What occurs during a recombination event? what is it? and WHAT MAKES THE RECOMBINATION?
DNA = broken = double strand break
DNA crosses over and the 2 are ligated together
Rec A makes the cross over
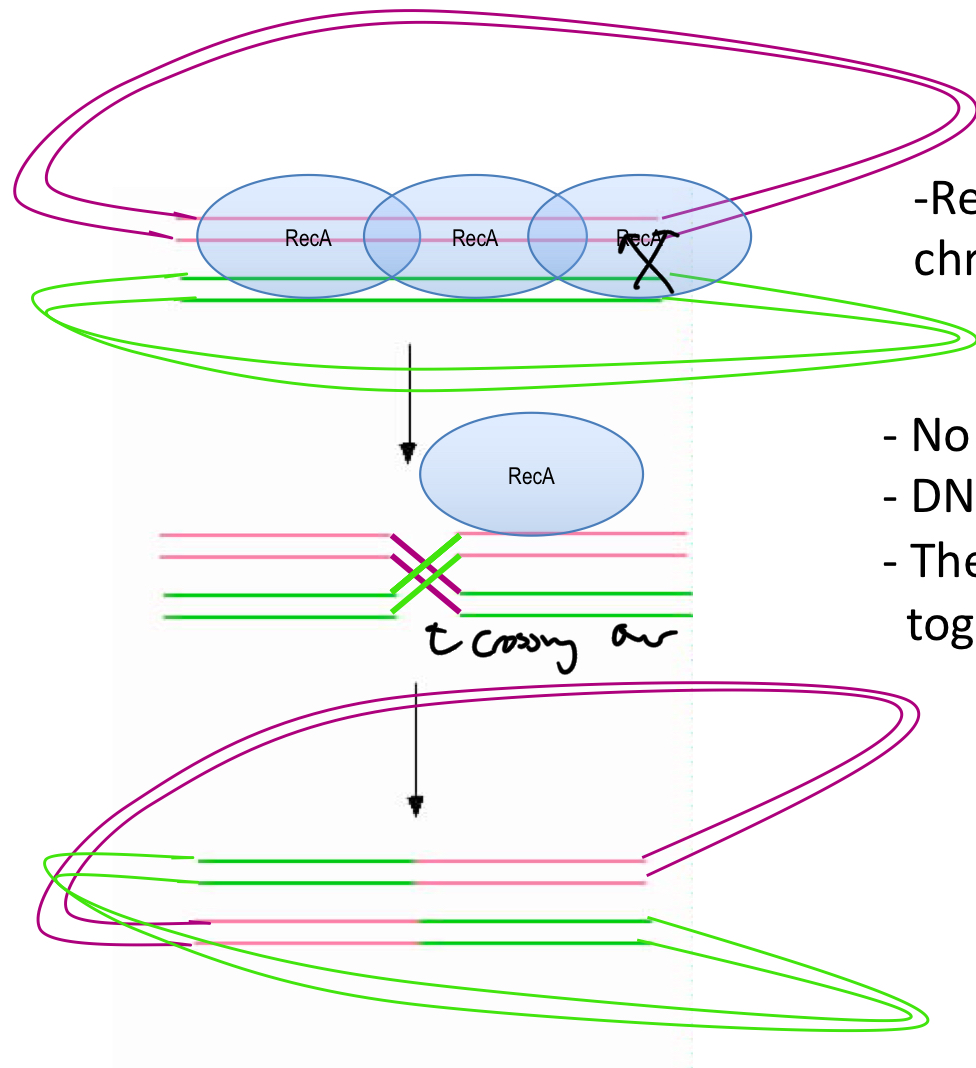
****Is any DNA added or lost during this process?
No
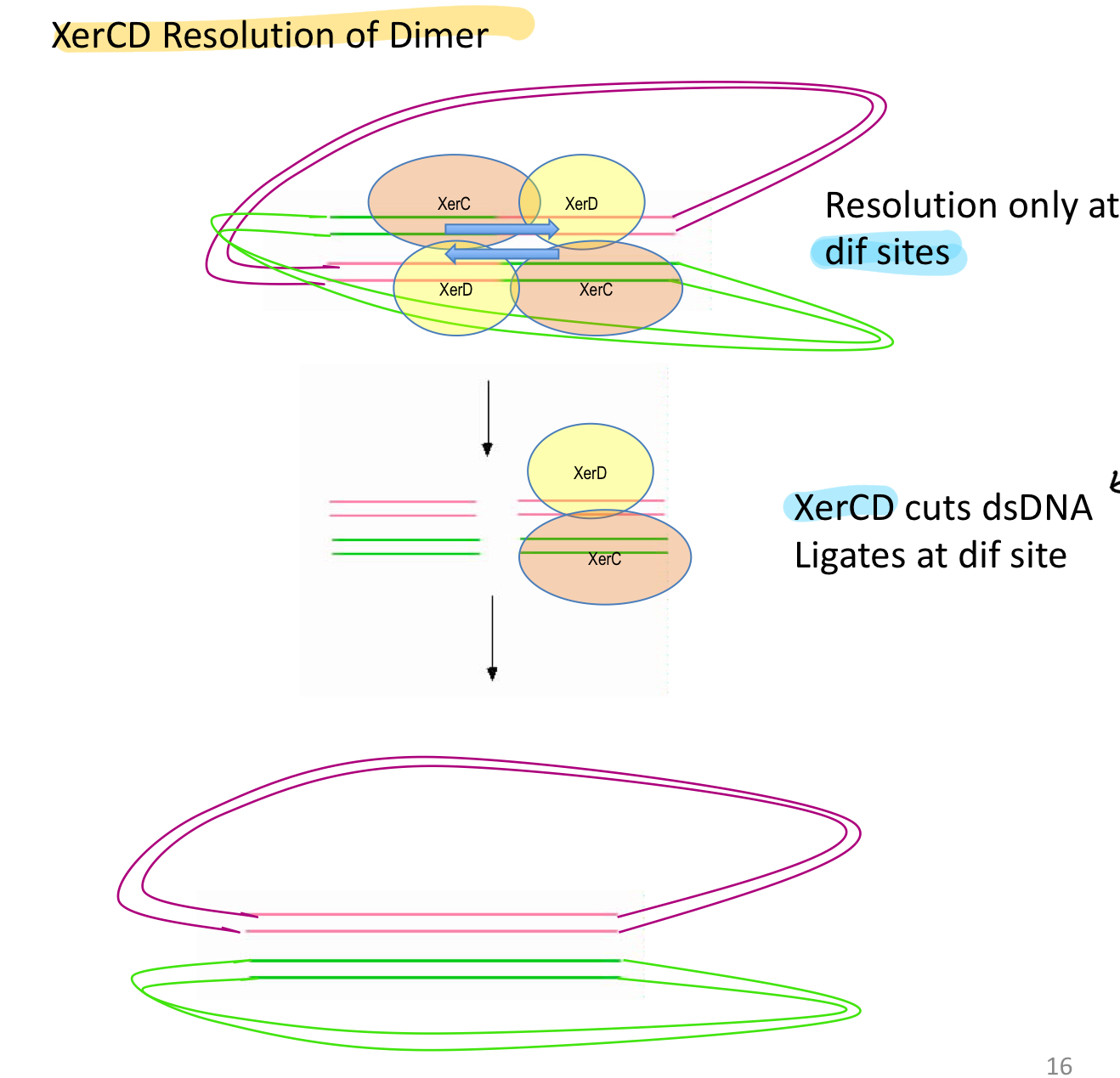
****What is the way Xer can resolve dimers?
Resolution by Xer CD
Xer Site-specific recombinase system
*****Explain how Xer CD works
Xer C + D cut dsDNA at specific dif sites —> REVERSE CROSSOVER = Ligates at dif sites
= 2 separate chromosomes instead of 1 dimer
****explain how Xer Site-specific Recombinase system works
Xer CD binds to Dif sites near ter
occurs when there are 2 copies of dif sites present on DNA
Interact with FtsK motor that channels dimer back + forth until all KOPS sequences = aligned in the right direction in each DNA + Dif site = in middle/ DNA evenly split
Xer CD = cut + ligate at dif site into 2 separate strands
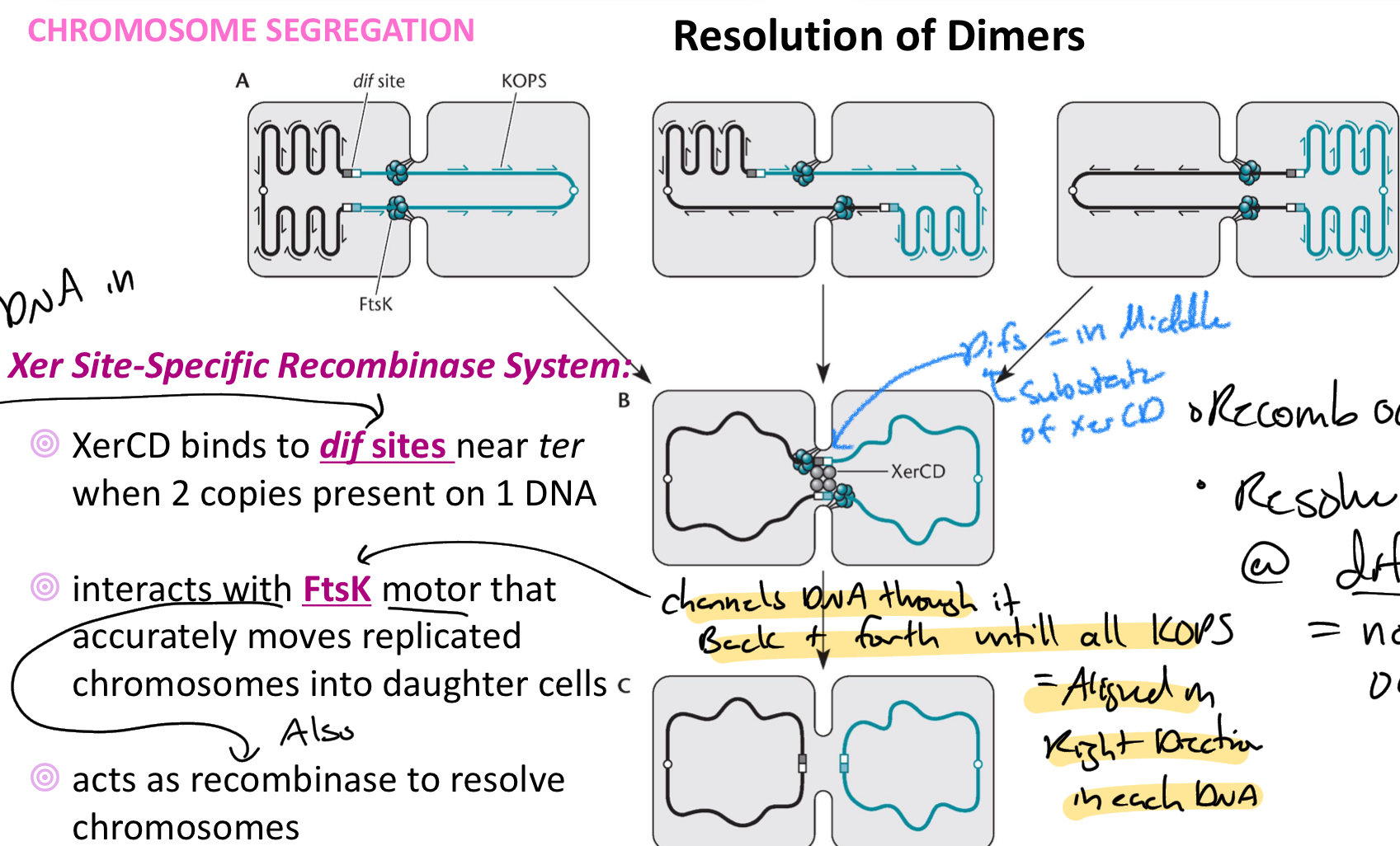
****What is the Kops sequence?
Goes in opposite direction in each DNA
1 side of KOPS = 1 diff site + the other side of KOPS = other diff site
Helps the Ftsk recognize which way to pump/channel the DNA + when they have evenly split the DNA between the 2 cells.
****Does the resolution of Dimers always occur where the recombination occurred?
NO
recombination occurs anywhere BUT Resolve = ALWAYS AT dif Sites
****What is FtsK? What role does it play in chromosome segregation? How does it do its role?
a CELL division protein
In chromosome segregation it acts as a DNA TRANSLOCASE that moves dif to septum
Uses KOPS (FtsK-oreinting polar sequence) to know which way to move the chromosome
*****Where is FtsK located?
At the DIVISION SEPTUM
anchored to CELL WALL AT SEPTUM
***What is a Catenanes?
Tangling of 2 separate circles of daughter DNA + formed during DNA replication
Complication #3 of chromosome segregation
*****How are Catenanes resolved?
Topoisomerase IV
****Of the 2 types of Topoisomerase, what type is IV?
TYPE 2
= Cuts BOTH STRANDS of one DNA + passes the other DNA through
*****What is Topoisomerase also in association with?
FtsK
*****How are Catenanes FORMED during DNA rep?
Positive supercoiling ahead of the fork
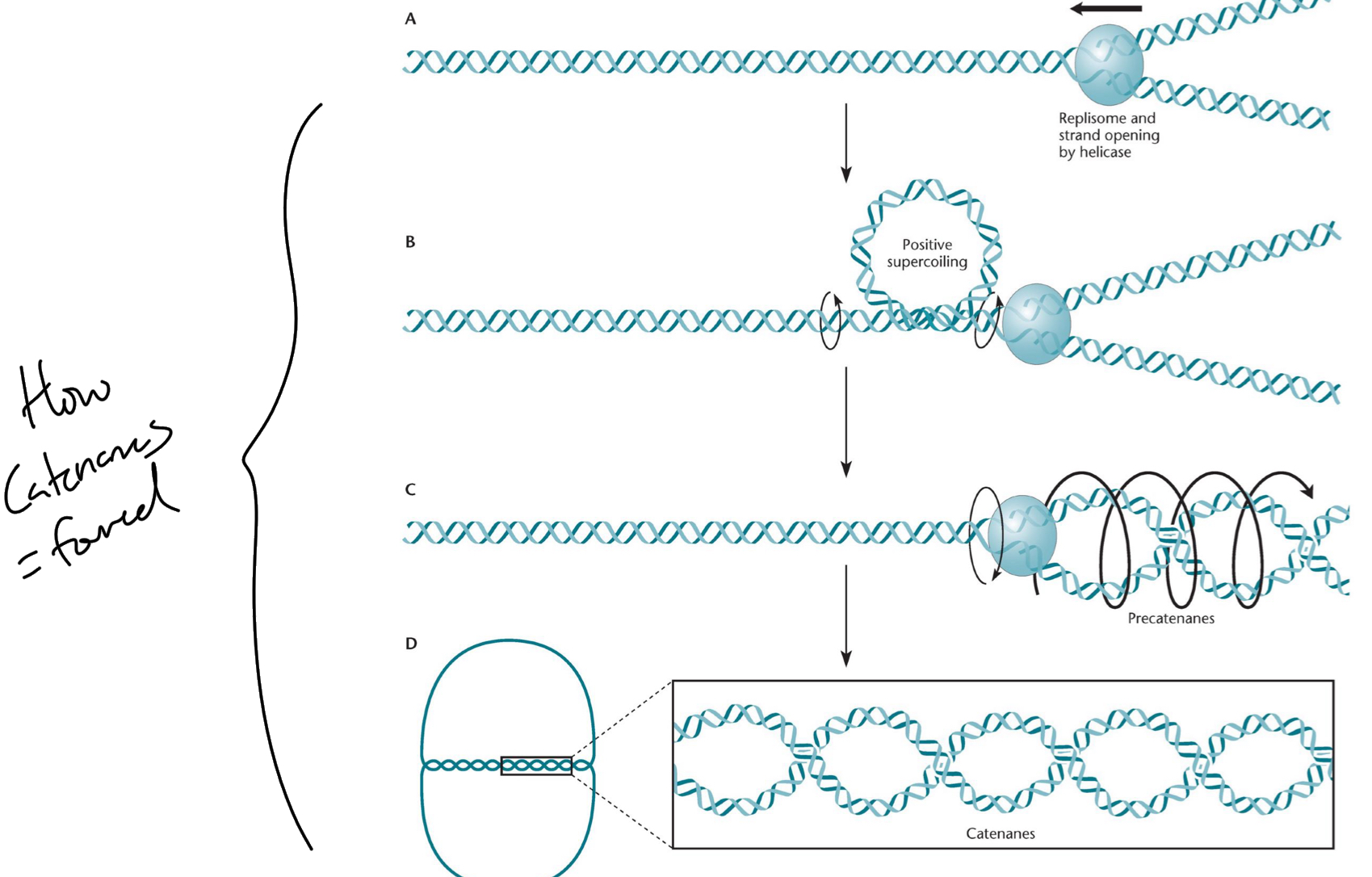
****What is the process of resolving catenanes called?
decatenation
****How does FtsK + Topoissomerase IV work to resolve catenanes?
Similar mechanism to Xer
THREAD DNA back + Forth until TANGLE = ALIGNED in the Middle/SEPTUM
Topoisomerase = Cut 1 strand + thread other through
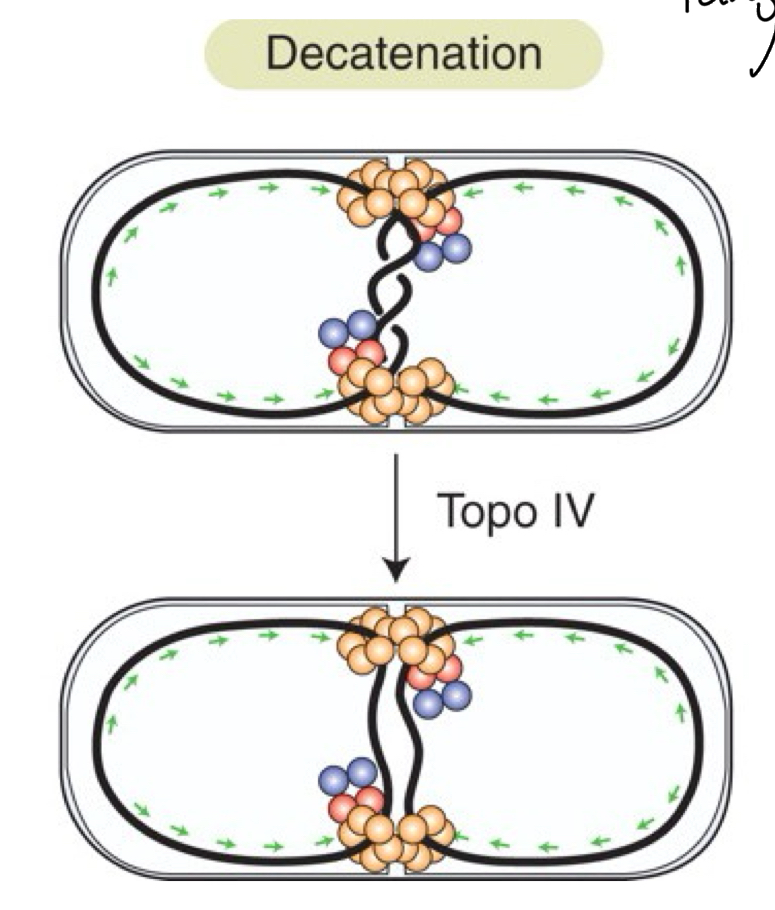
Does the decatenation process use KOPS?
NO
instead aligns the tangle with the middle
*****Finish the Sentence: Topoisomerase IV is active based on _____?
Based on DNA strain
****What is Condensin? What does it Do?
Protein that works with TOPO IV to condense DNA after decatenation.
Condense newly replicated chromosomes' so there is less change of tangling
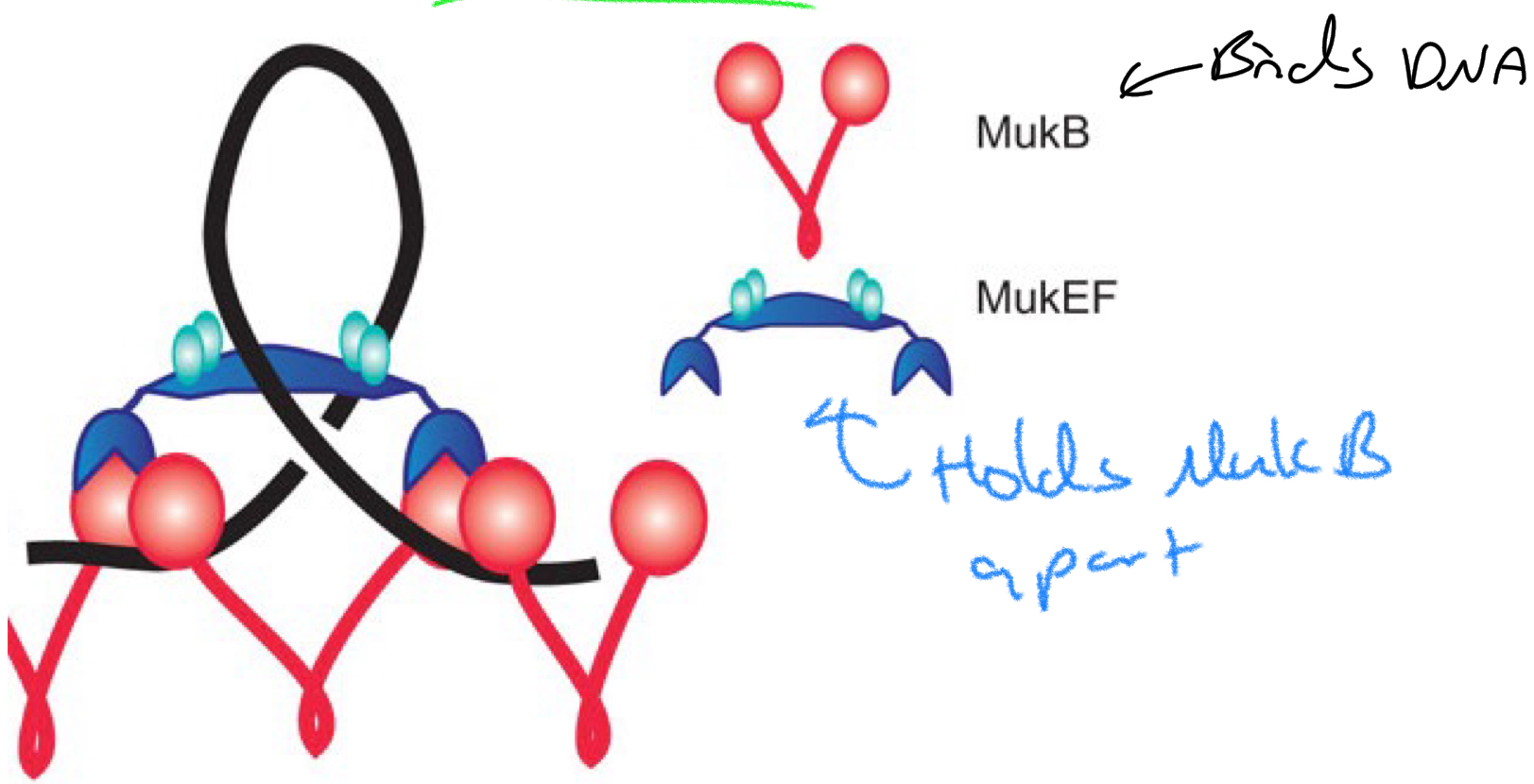
*****What is the condensin protein in E.Coli?
MukB
*****What does MukB look like + how does it work?
Dumbbell shaped proteins
Bind DNA + allows binding of Muk EF to hold it apart in large loops
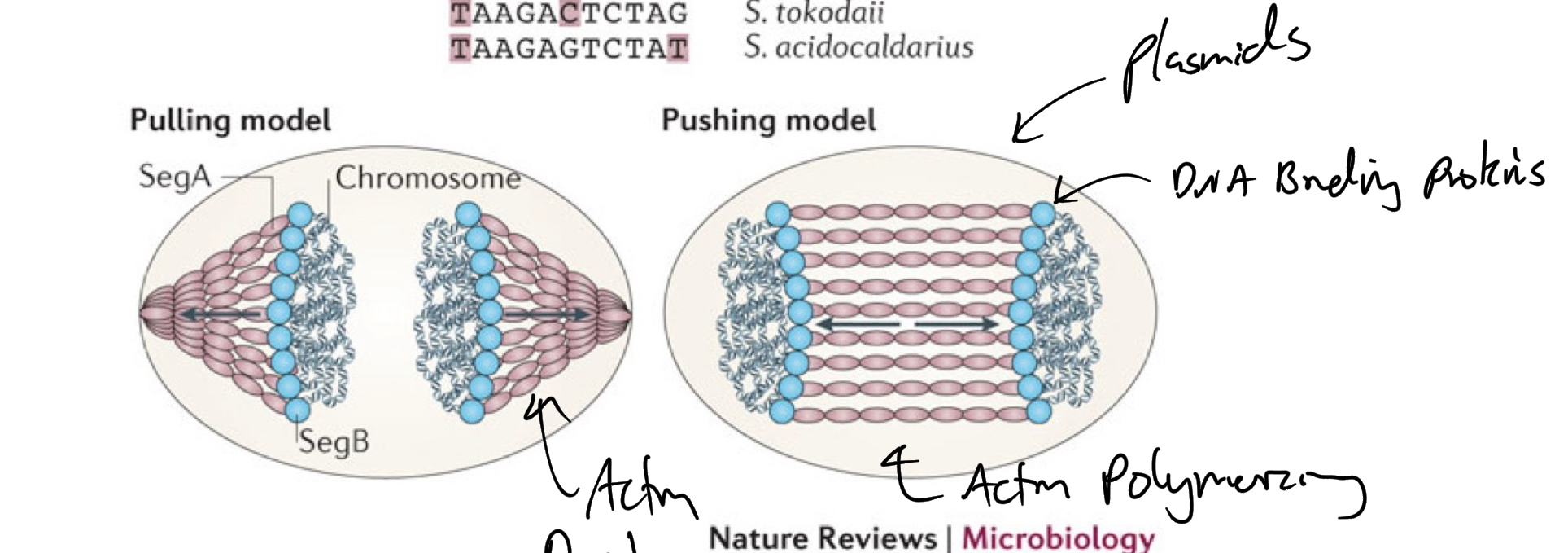
*****What are the 2 types of Chromosome Partitioning?
Pulling mechanism
Pushing Mechanism
Fill in the blank: Partitioning systems resemble those of ______ and vary between bacteria.
Resemble plasmids
*****What PROTEINS are involved in the process of Partitioning for the pushing + pulling mechanisms? What general way do they achieve this?
Par proteins (actin like)
Push by polymerization
Pull by Depolymerization
*****What are the 2 MECHANISMS that work together + are responsible for Coordinating cell division + chromosome partitioning How do Cells divide in the middle + ensure each daughter cell gets 1 chromosome?
Min proteins
Nucleoid Occlusion
*****1. How do Min proteins work to coordinate cell division?
Ensures septum formation only occurs in the MIDDLE
Septum cannot form where there is min
works either by oscillation mechanism or by polar localization
******2. How does Nucleoid occlusion work to coordinate cell division?
Nucleoid occlusion proteins bind DNA + prevent FtsZ division ring/septum
Prevents cell division from cutting chromosome in half before DNA is finished replicating
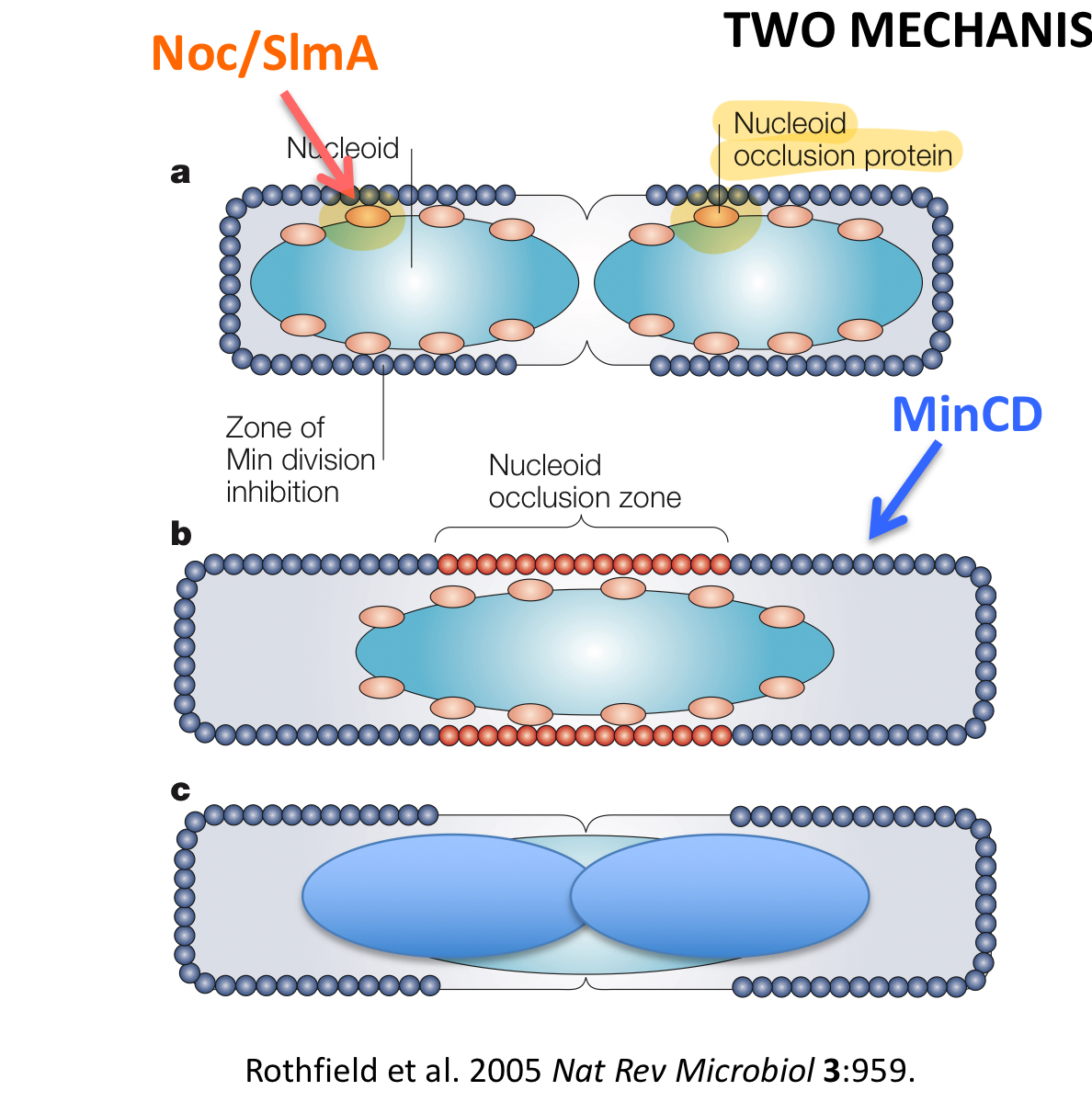
*****What are the 2 nucleoid occlusion proteins talked about in class?
Noc
SImA
****In which ORGANISMS do theses 2 proteins each occur?
Noc = B. Subtilis (GRAM +)
SImA = E.coli = (GRAM —)
*****What are the 3 MECHANISMS responsible for controlling the timing of chromosome replication? How does it a cell know when to replicate?
Cell Mass
DnaA binding sites
Hemi-methylation + sequestration
******Which scientists ran an experiment that determined CELL MASS was a mechanism of control for replication?
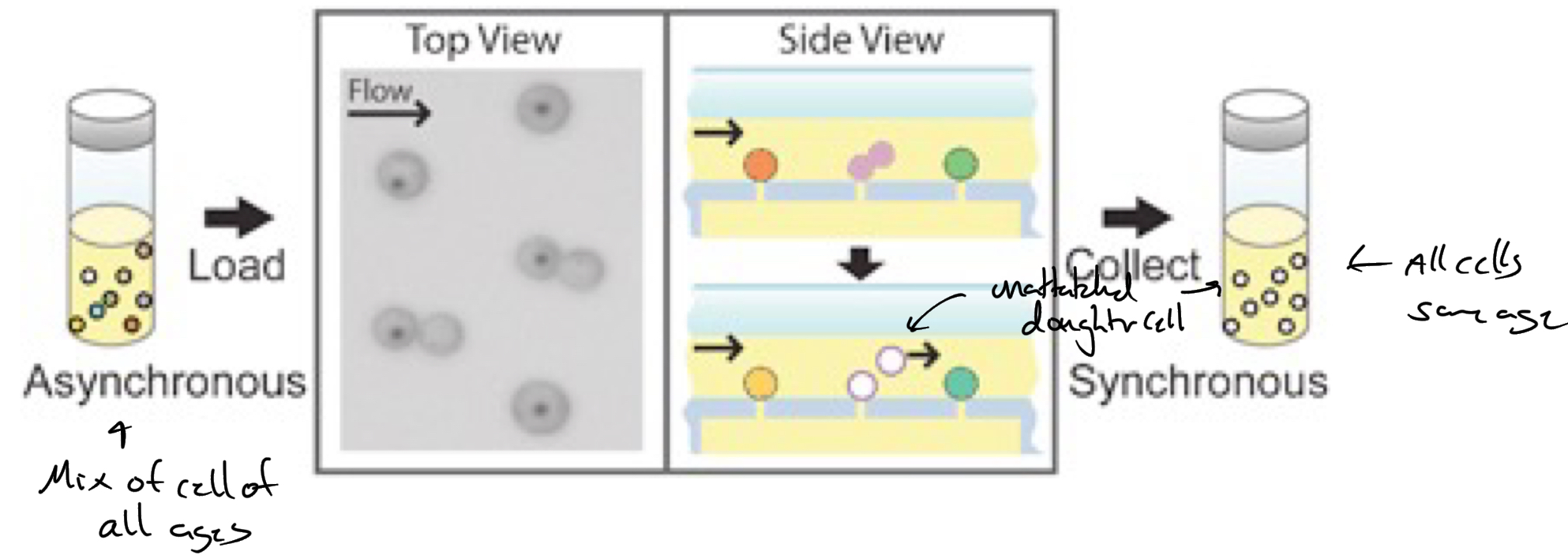
Helmstetter + cooper
baby machine experiment
*****How did the baby machine experiment work?
Measure DNA conenta @ different stages of cell cycle
Attachment of “mother” cells
sticking them onto a membrane filter
SYNCHRONIZE BACTERIA
Newly divided cell = not attached + WASHED into the flowing medium.
These newly released cells were newborns (age = 0 minutes).
Collecting synchronized cells
The flow system continuously washed away “babies,” giving a pure, synchronized population of cells all starting at the same cell cycle point.
Measure radioactive DNA content to determine how much chromosome replicated at time X
********** What did Helmstetter + cooper DIVSOVER? What 2 conclusions did they make?
Time BETWEEN initiation of replication differed
Time taken for replication events + cell divisions events to be completed = CONSTANT
******What affected the time BETWEEN initiation of replication?
Nutrient availability + RATE OF GROWTH
high availability = faster growth = faster imitation of replication = decrease time between
******What does the 2 conclusion from the baby machine experiment mean/ tell us?
More than one replication round may be initiated in the same cell when growth rates are fast

******What are the 2 WAYS DnaA binding sites affect Timing of replication?
Cellular Concentration of Dna A
ATP vs. ADP bound state of Dna A
*****how does CELLULAR CONCENTRATION of DnaA influence rate of replication?
Replication = Number of oriC sites DOUBLED
DnaA binding = slower
Other DnaA binding sites also compete with oriC for binding DnaA
= Less DnaA available to bind to Duplicated OriC site to initiate replication
*****how does ATP vs. ADP bound state of DnaA of DnaA influence rate of replication?
Only ATP-Bound DnaA can bind oriC DnaA boxes (I, sigma + DUE) to initiate replication
DnaN = causes DnaA to HYDROLYZE bound ATP = no ATP-bound not available = Inactivate initiation until there is more ATP
inactivate DnaA-ATP so oriC doesn’t fire again.
*****What is DnaN + what does it do?
AKA = Beta sliding-clamp = ATPase
Hydrolyses ATP bound to DnaA to prevent immediate replication
***What are the 3 things Replication depends on?
DnaA
# of DnaA binding sites
[DnaA-ATP]
How can replication be initiated faster?
High concentration of ATP = can replenish ATP faster of DnaA
***********how does Hemi-methylation + sequestration influence rate of replication?
DNA = methylated
After replication DNA = HEMIMETHYLATED (only one strand methylated)
SeqA binds to oriC in un-methylated areas = blocks binding of DnaA-ATP
Bound SeqA = drawn to cell membrane + hidden from methylation enzyme
Once found + methylated, SeqA falls off of oriC = DnaA-ATP can bind
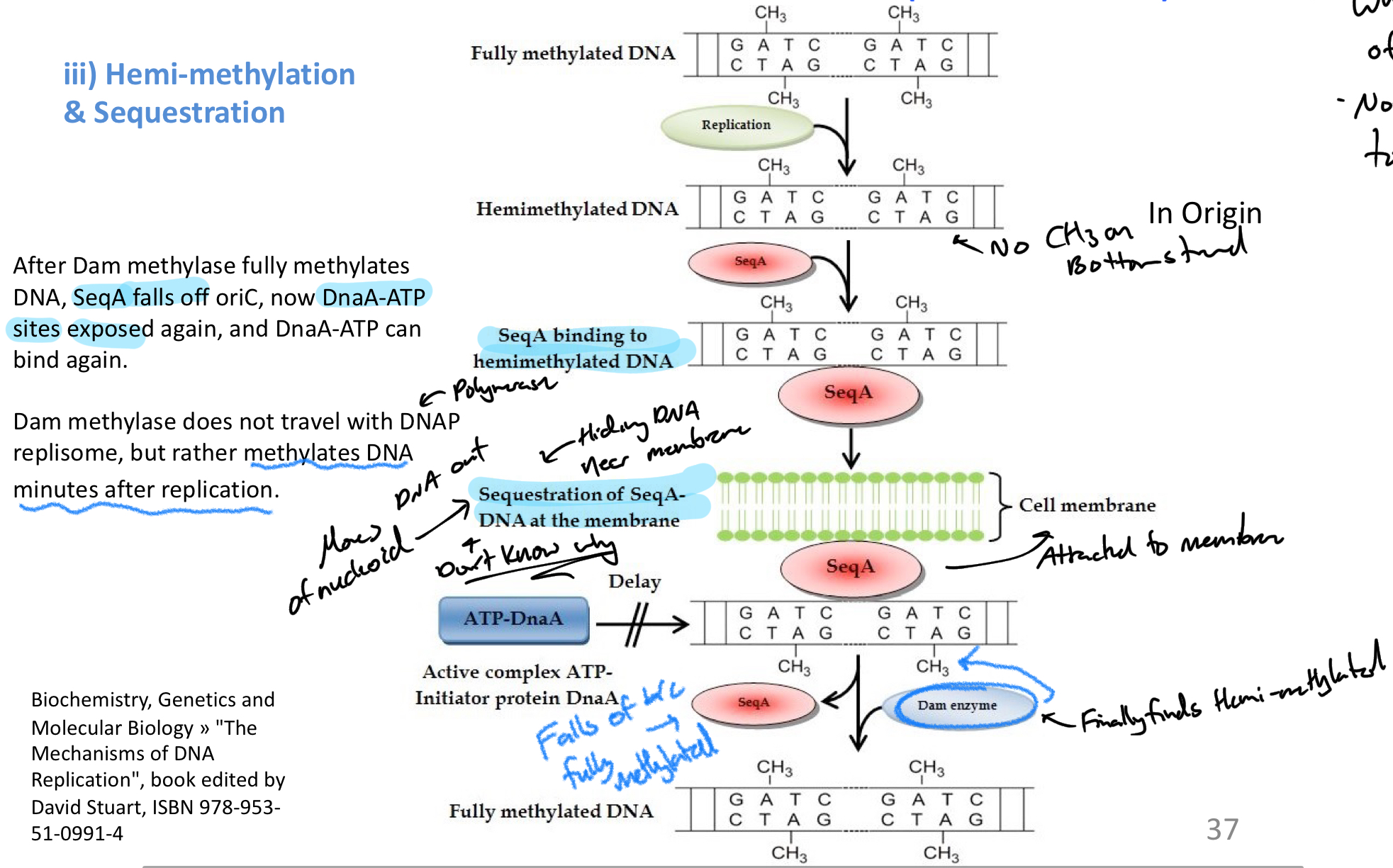
*****What ENZYME methylated DNA in E.Coli?
Dam methylase
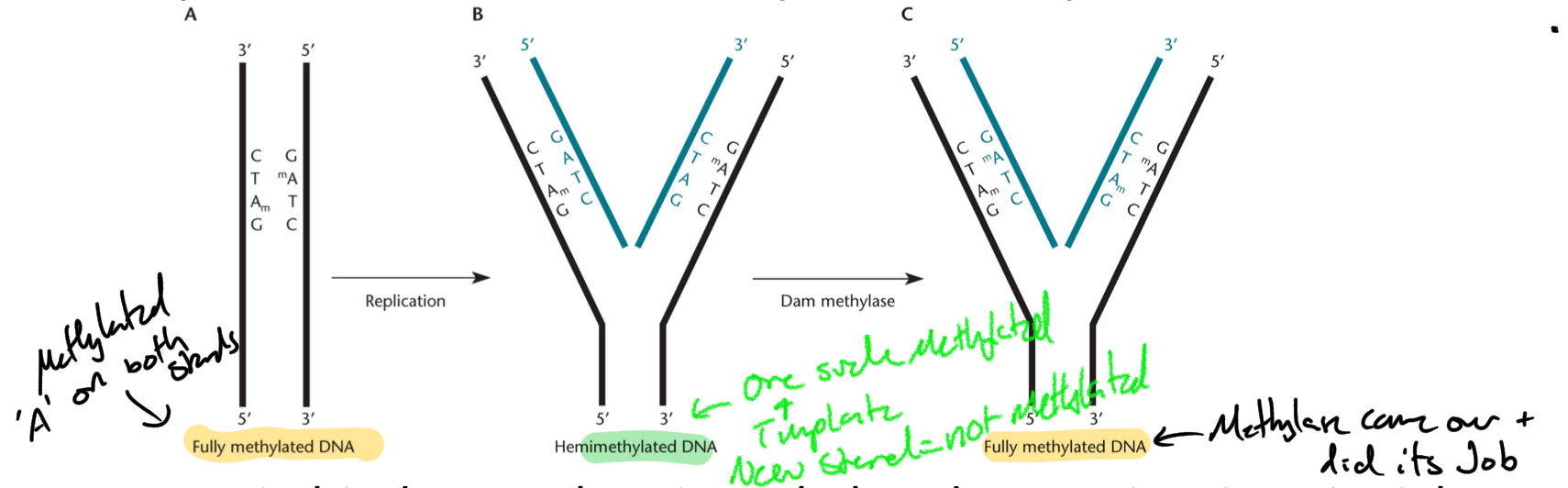
****Where is E.coli methylated? What specific sequence?
5‘- GATC -3’
****How many GATC sites are there in oriC in which SeqA can bind to the unmethylated version?
11
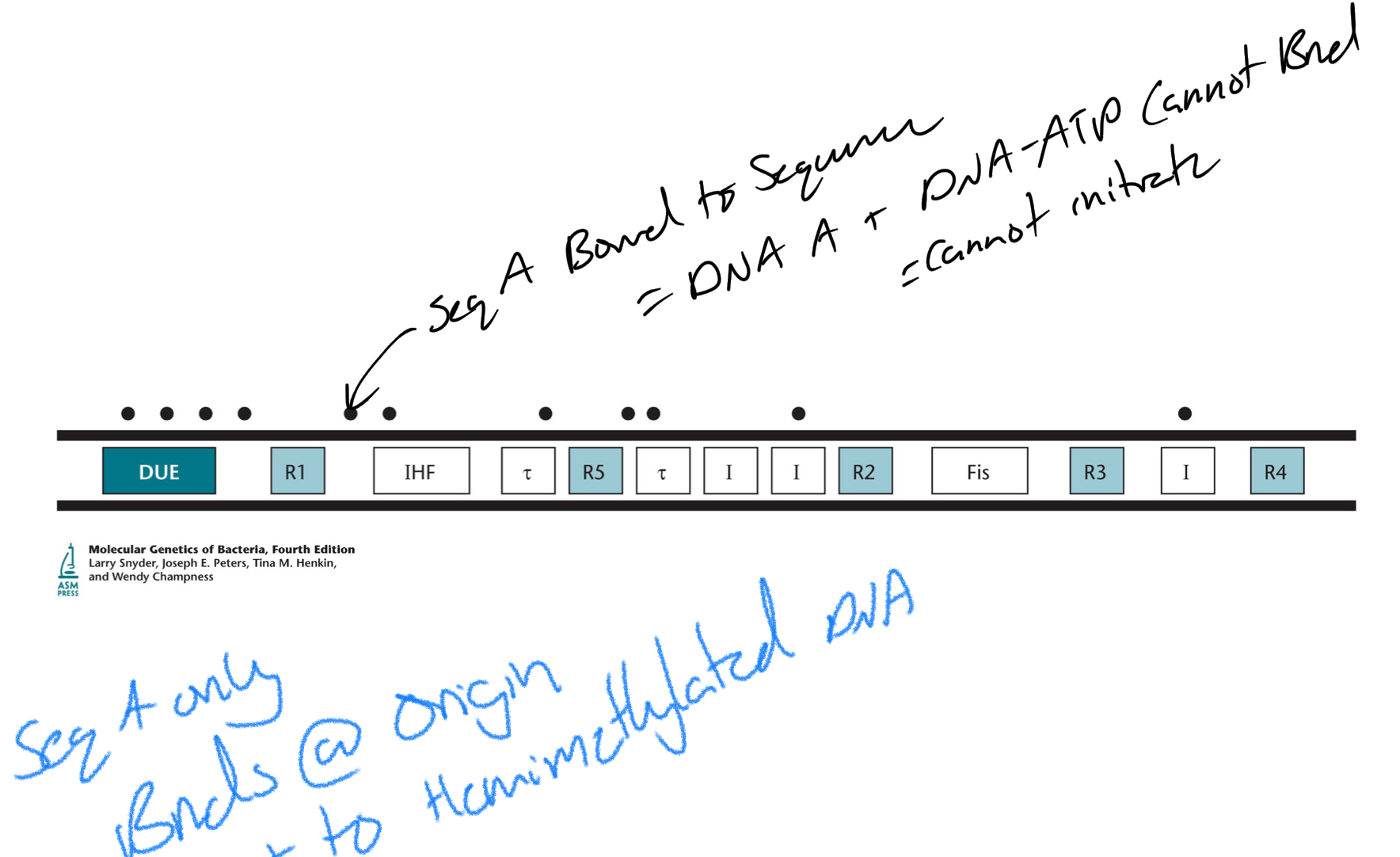
*****What is the purpose of all the replication timing mechanisms?
Not start replication too quickly
ensure there is enough E
Prevent DNA rep from going out of hand