BIO130 first half
1/49
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
50 Terms
Week 1: cellular diversity: Cell Theory
The cell is the basic unit of life
All organisms 1 or more cells
Cells arise from pre-exsisting cells
Prokarotic cell
No membrane-bound organelles
Smaller than eukaryotes
Less DNA
DNA is compartmentalized but no membrane —> nucleoid
Eukaryotic cell
Nucleus
Membrane-bound organelles
Larger and more complex
Origins of mitochondria
Entangle-engulf-endogenize (E3 ) model
Ancient anaerobic archeal cell and ancient aerobic bacterium
started as ectosymbiote (outside)
then engulfed as endosymbiote
then bacteria membrane broken
(other model show predatory mechanism)
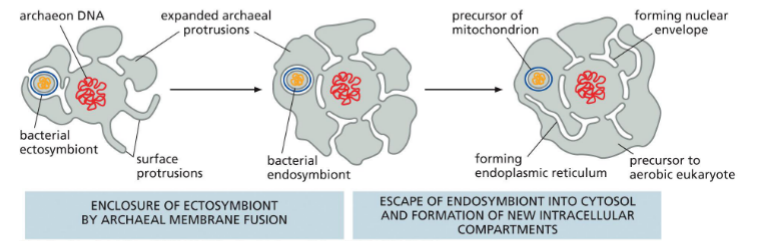
Ancient cell folding protusions
Give way for nuclear envelope and endoplasmic reticulum
Origins of eukaryotes graph
mitochondria first —→ eukaryotes
chloroplast second —→ plants
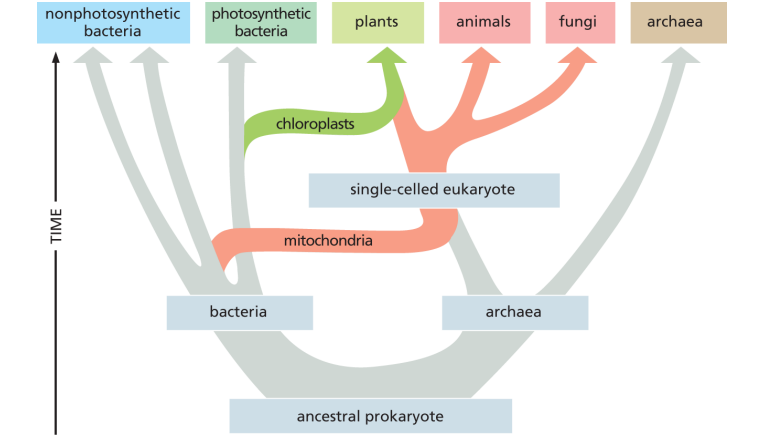
Endosymbiont hypothesis for mitochondria and chloroplasts — evidence
Both have remnants of own genome which resemble modern prokaryotes
Both have kept some of own protein and DNA synthesis components, also resemble prokaryotes
Membranes are similar to prokaryotes and derived from bacteria ancestor
Model organisms and Humans — general attributes of model organisms
Fast development and short life cycles
Small reproductive (adult) size
Readily available
Tractability (manipulation or modification)
Understandable genetics
Examples of model organisms
Ecoli
Brewer’s yeast — simple eukaryote
Arabidopsis — plant
Nematode, drosophila, zebrafish, mice — animals
Lec 2: The Central Dogma of Molecular Biology
Information flow is always in one direction
DNA (transcription) —→ RNA (translation) —→ Protein
.
Refined:
messengerRNA: translation for protein
transferRNA: transport amino acids
rRNA: heart of ribosome, breaking and forming of bonds
Antiparallel and genetic code
DNA, RNA, and proteins are synthesized as linear chains of info with intrinsic directtion
RNA translating to amino acid is universal through the genetic code
What are nucleic acids?
Genetic material in a cell
DNA & RNA
Three parts of a nucleotide
Pentose sugar
Phosphate group (1, 2, or 3)
Nitrogenous base
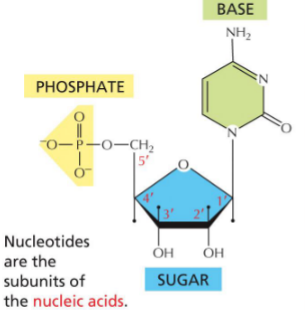
Nucleotide bases
Pyrimidine
Cytosine
Thymine
Uracil
Purines
Adenine
Guanine
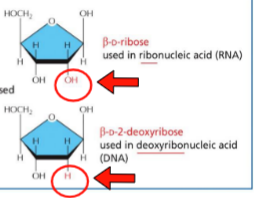
Differences between DNA and RNA
RNA:
ribose
OH on 2’ carbon
uracil
DNA:
deoxyribose
H on 2’ carbon
thymine (extra methyl)
Differences between bases
Nucleic acid nomenclature
Nucleoside: sugar + base
Nucleotide: sugar + base + 1, 2, 3 phosphate
Nucleoside monophosphate
Nucleoside diphosphate
Nucleoside triphosphate
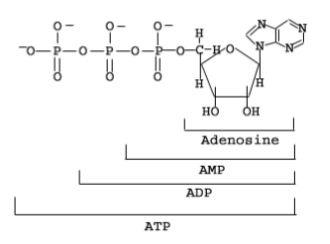
Nucleic acid chains
DNA is synthesized from deoxyribonucleoside triphosphates (dNTPs)
RNA is synthesized from ribonucleoside triphosphates (NTPs)
Nucleotides linked by phosphodiester bonds
Molecular interactions
Electrostatic attrations (charges attract)
Hydrogen bonds
Van der waals attractions
Hydrophobic force
(Individually weak, sum to be strong)
Three forces that keep DNA strands together
H-bonds (base pairing, G-C stronger cause 3 bonds)
Hydrophobic interactions (phosphate backbone hydrophylic, bases hydrophobic
Van der waals attractions
DNA structure
DNA will naturally come together, energetically favourable
Proteins can recognize and make contact with specfic sequences in major & minor grooves
Separating DNA strands
DNA can unravel and return to double helix
use energy or enzymes to denature
useful for replication, transcription, or PCR
Lec 3: intro to protein structure
Quaternary: more than 1 polypeptide chain, subunits
Multiprotein complexes: many chains and subunits, machine
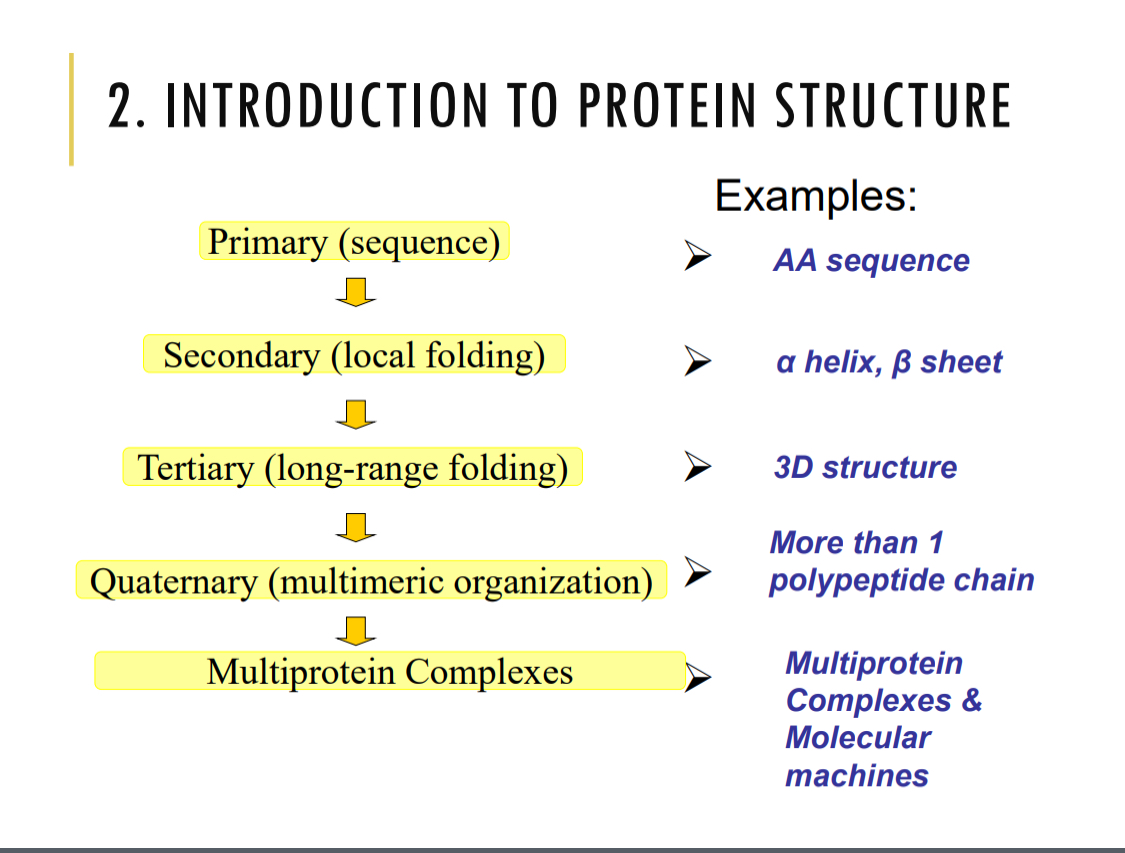
Amino acid structure
proteins composed of amino acids
side-chain/R group is variable and determines the type of amino acid
three major categories
acidic ( - charge)
basic ( + charge)
uncharged polar
nonpolar
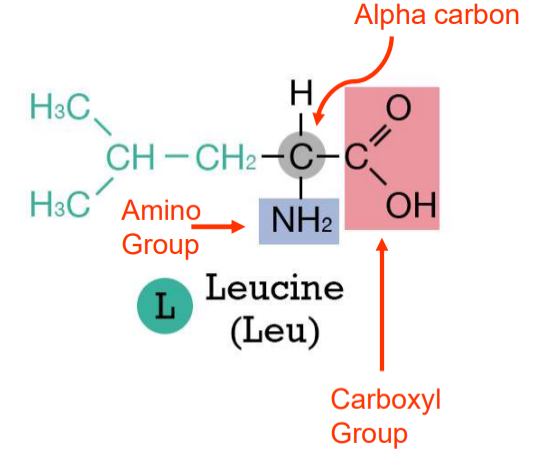
classification system for amino acids
half are polar
5 charged polar
5 uncharged polar
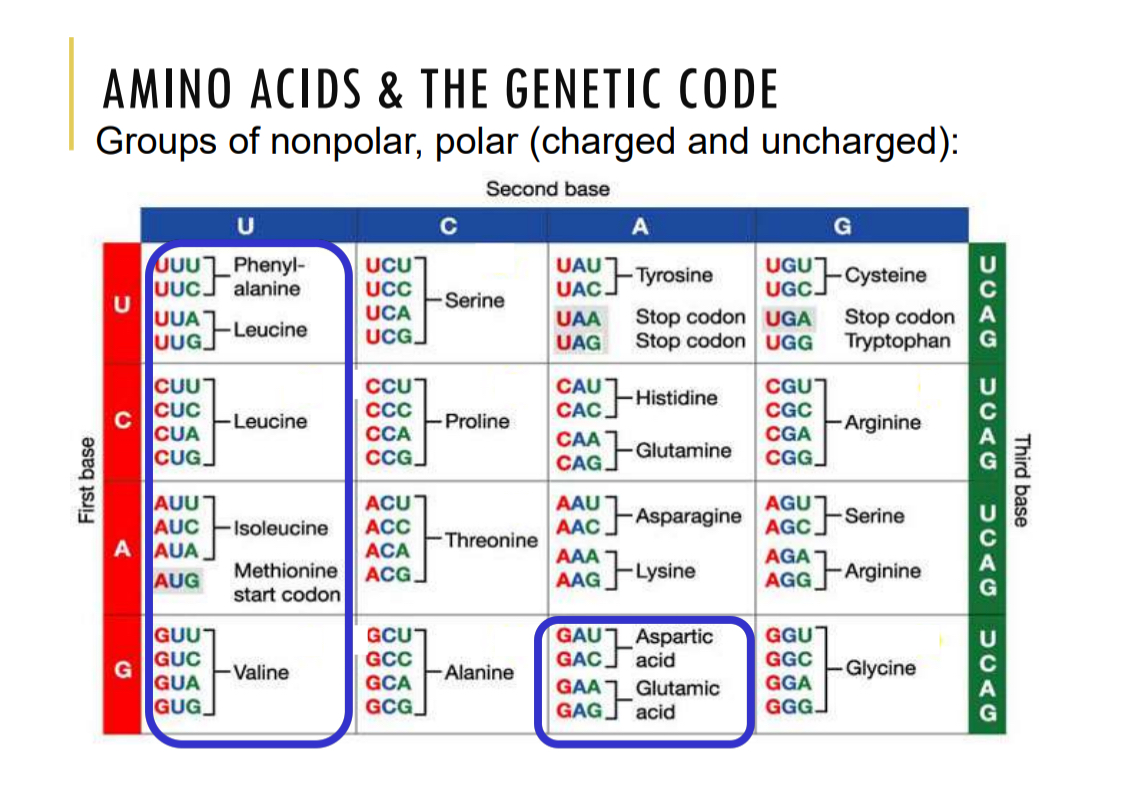
Amino acids and the genetic code
AUG — start codon methione
UAA, UAG, UGA — stop codon
Degenerative code: more codons than AA, more than one can code for the same AA
Flexibility: similar codons for similar AA, can tolerate mutations better
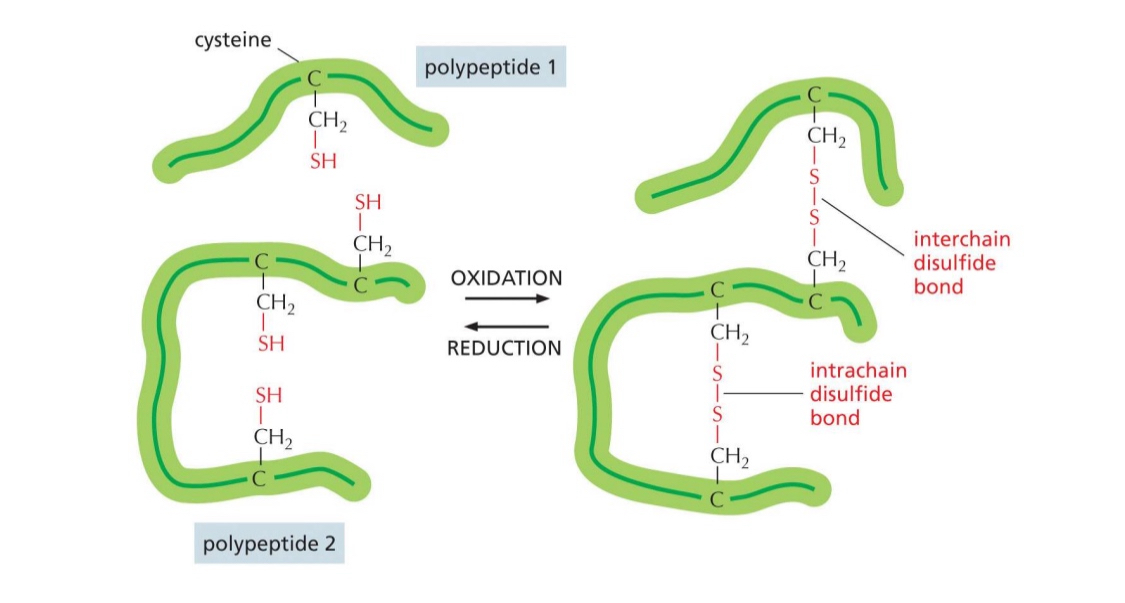
Unique amino acid: cysteine
can form disulfide bonds — (oxidation form, reduction break)
both interchain and intrachain
covalent bond creates stability
“staple”
often used in structural proteins
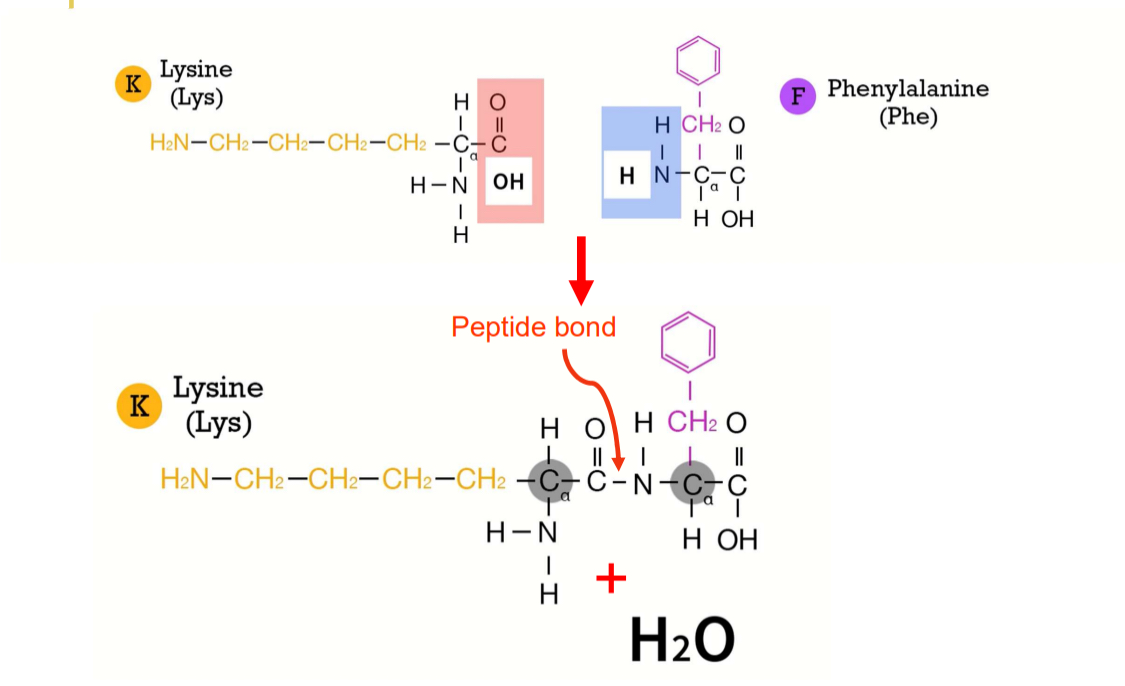
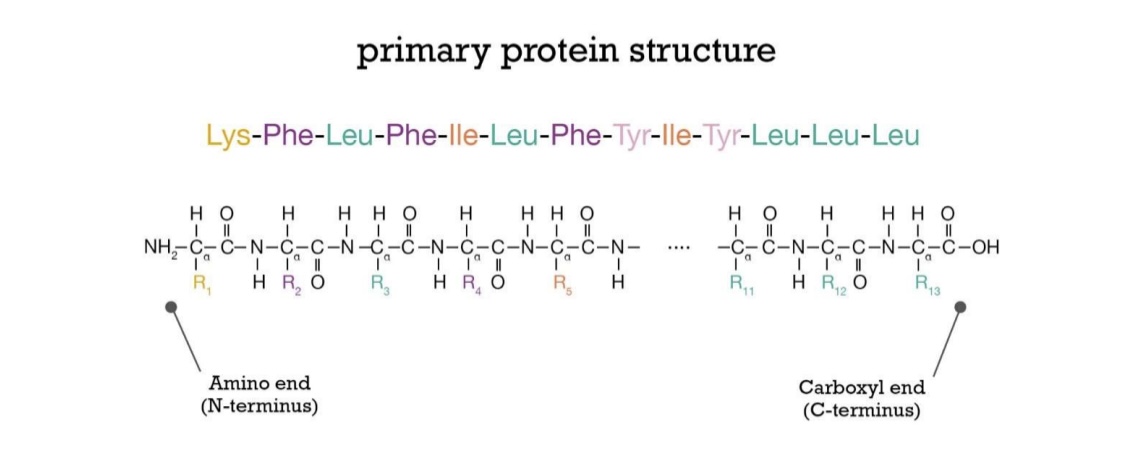
Primary structure: Peptide bonds
catalyzed by ribosome
peptide backbone of C-C-N-C-C-N
Polarity: always grow in the same direction, starting at N-terminus
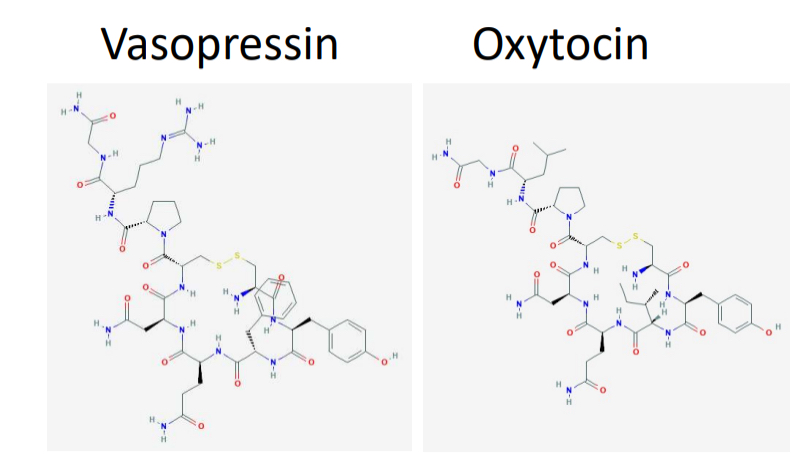
Differences in primary AA sequence matter - vasopressin example
both vasopressin and oxytocin are 9 AA long
both are identical except at two locations
vasopressin controls urine production
oxytocin involved in birth, lactation, and pair bonding
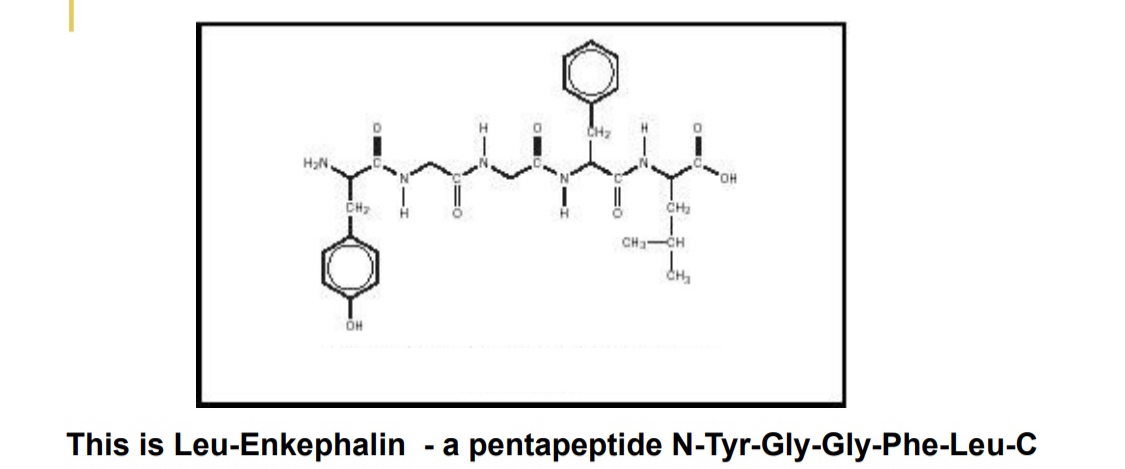
Order of AA is important too - Leu-enkephalin
natural opioid
the opposite order of AA has no pharmalogical effects
the amine-carboxyl orientation essential to function
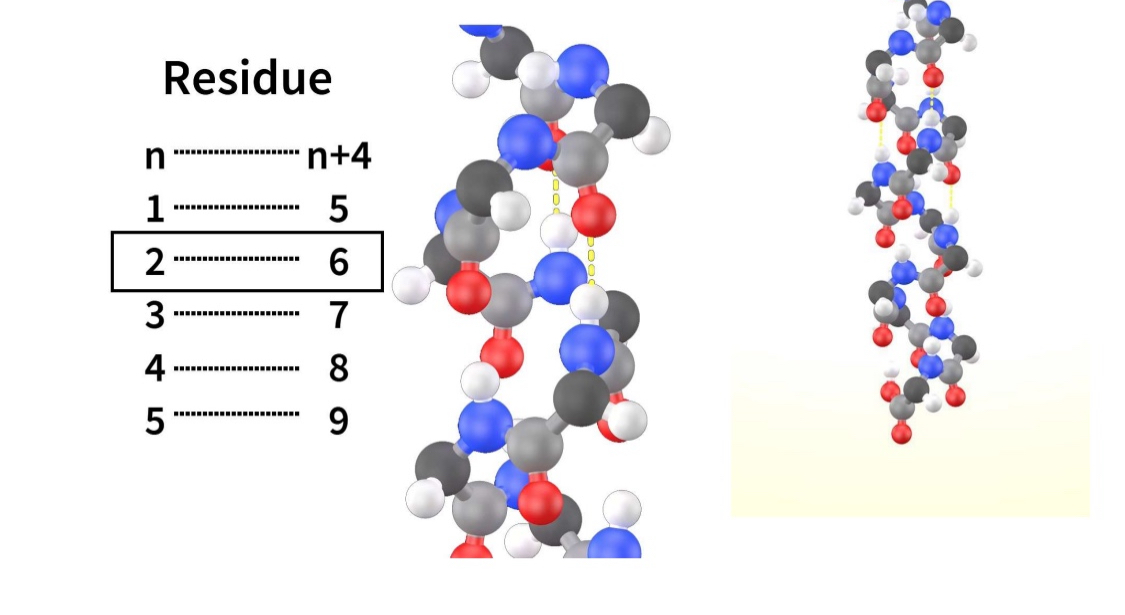
Secondary structure: Alpha-helix
Forms independantly of side chains
carboxyl h-bonds with amino of AA 4 after
n — n+4
(helical structures are common in biology because they are stable
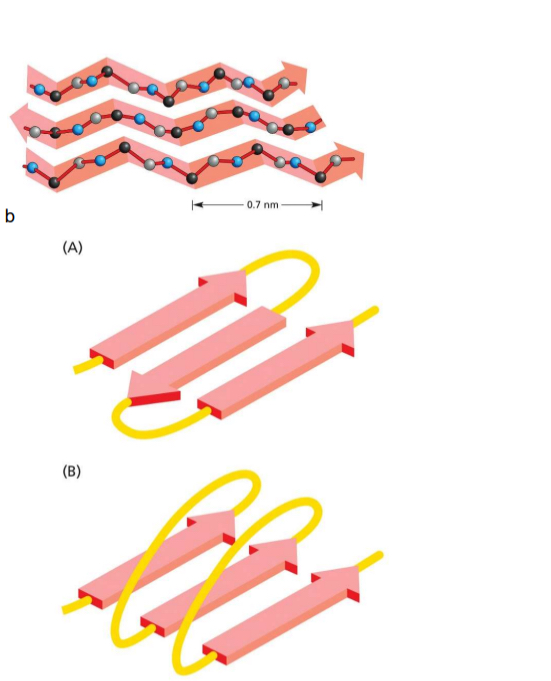
Beta sheet
Forms independtly to R groups (but they alternatively point up and down, interactions)
H-bond of carbonyl (C=O) with amide hydrogen (N-H) of neighbouring strand
typically contain 4-5 strands but can have more
can be antiparallel or parallel
anti only needs small sequence between
parallel needs more
Counting polypeptide chains
Always count from N-terminus (amino end)
Coiled coil
Alpha helices twisting together
Only form with amphipathic — protein with both hydrophobic and hydrophilic parts
repeating hydrophobic molecule every 4 peptide bonds — hydrophobic stripe
2 helices will wrap together, push hydrophobic parts into middle
very stable and strong
keratin in hair
myosin motor proteins

Amyloid structure
Beta sheets stacked together
Misfolded proteins can form amyloid structure — neurodegenerative diseases
prions: converts properly folded molecules
Lec 4: Tertiary structure
Overall 3D structure of a protein
Held together by:
hydrophobic forces
non-covalent bonds
covalent disulfide bonds
Hydrophobic force
non-polar AA in interior of folds
polar AA on exterior
Tertiary structure — continued
Proteins fold into conformation that is most energetically favourable — spontaneous
H-bonding in:
backbone/backbone
backbone/side chain
side chain/side chain
Chaperone proteins can also help make process more efficient and reliable in living cells
misfolded proteins cant function
Tertiary structure can have large variety of shapes
globular, filament, etc
But few of the possible chains will be useful
majority 50—2000 AA long
well-behaved, stable
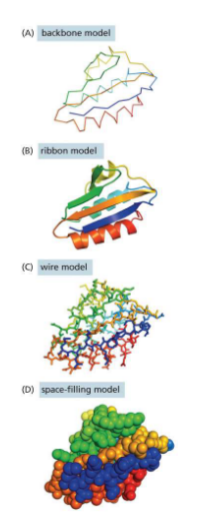
Models for proteins
Backbone model — only backbones
Ribbon model — shows folding
Wire model — shows positions of bonds
Space-filling model — contour map
Protein domains
Regions of proteins that have specialized functions
single polypeptide
each domain has own tertiary structure and function semi-independently
.
eukaryotic proteins often have 2 or more
connected by intrinsically disordered sequences (flexible regions)
domains are important for the evolution of proteins
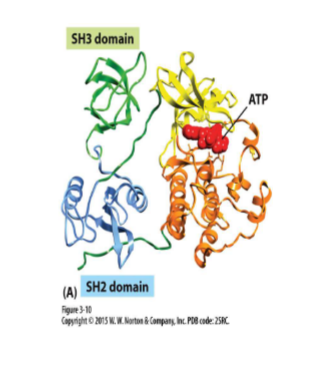
Protein domains — extra example: Src protein kinase
Kinase — phosphorylate proteins (changes activity)
Src protein kinase has 3 domains
SH2 and SH3 regulates kinase
Protein families
Way to organize proteins — a protein can belong in more than one
Similar AA sequences and tertiary structures
Members have evolved different functions
Most proteins belong to families with similar structural domains
Quaternary structure
More than1 polypeptide chain
not all proteins
subunit = separate polypeptide
can get really big
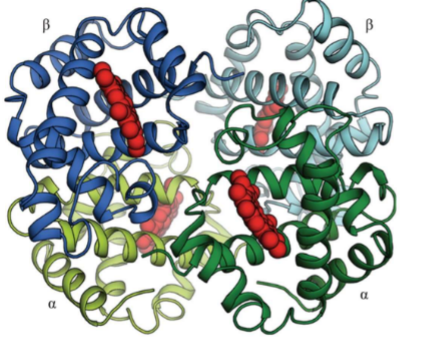
Quaternary structure example: Hemoglobin
Each hemoglobin has 4 subunits (2 alpha, 2 beta)
Sickle cell anemia is caused by mutation in beta subunit
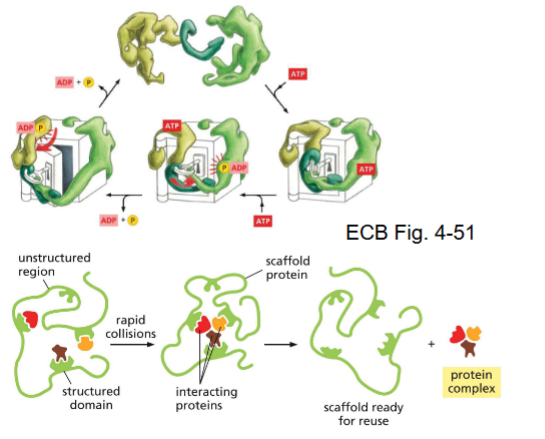
Multiprotein complexes and molecular machines
Can be:
many identical subunits (actin filaments)
mixtures of proteins and DNA/RNA (ribosomes)
dynamic assemblies of proteins to form machines (DNA replication)
Multiprotein complexes and molecular machines
conformational changes
perform job
often need ATP
.
scaffold proteins
binds proteins together
How are proteins studied?
Past
purify proteins
electrophoresis & affinity chromatography
.
Now
Mass spectrometry
sequenced many genomes
find mass and match to predictions
discover precise 3D structure with other techniques
can also use AI to predict structure using only polypeptide
Protein separation
separate using size, shape, charge, hydrophobicity
Proteomics
Large scale study of proteins
structure
interactions
abundance and turnover
location