MBIO 2020 / Topic 1: Nucleic Acids
1/27
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
28 Terms
briefly differentiate and or define these terms: gene, DNA, chromosome, genome, and plasmid
> gene = DNA segment
> DNA = macromolecule containing genes
> chromosome = package of DNA containing essential genes
> plasmid = piece of DNA containing non-essential genes
> genome = full set of chromosomes in organism
> what are the two key types of nucleic acids?
> what’s the difference between them in both structure and genetic function?
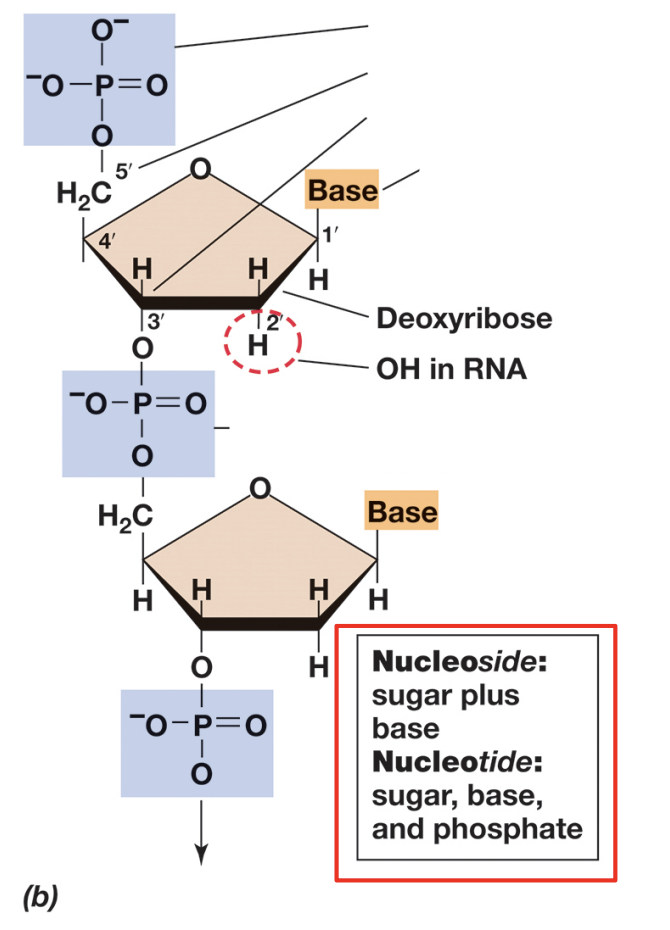
> deoxyribonucleic acid (DNA), which differentiates by carrying 2’ deoxy-D-Ribose (H on 2’).
> ribonucleic acid (RNA), which differentiates by carrying D-Ribose (OH on 2’).
> both carry genetic information in the sequence of nucleotide monomers, but RNA carries a copy of this genetic information (from the DNA) for protein synthesis
What are the three key components of a nucleotide?
What is a nucleoside?
A nitrogenous (nitrogen-containing) base.
A pentose sugar.
One, two, or three phosphates.
A nucleoside is just a nucleotide without the phosphate group.
What are the five possible bases for a nucleotide?
What are the two groups for these five bases? Why are they grouped as such?
The purine bases (adenine and guanine) are called purine bases because their parent compounds are purines.
The pyrimidine bases (cytosine, thymine for DNA, and uracil for RNA) are called pyrimidine bases because their parent compounds are pyrimidines.
Through what molecular forces do bases pair and thus keeps the double helix held together?
What are the two key stabilizing features of the double helix?
Why are C:G or G:C pairs stronger than A:T or T:A pairs?
DNA is held together by hydrogen bonding between base pairs.
DNA is significantly stabilized by (1) the presence of metal cations that shield the negative charges of phosphate groups and (2) the base-stacking interactions of successive C:G or G:C pairs.
G:C pairs have three hydrogen bonds, while A:T pairs only have two.
> NAs are made of nts, but what holds each nt together and in what way?
> how are nucleotide sequences usually written? 3’ to 5’ or 5’ to 3’?
> what is our backbone and side groups on NAs?
> nts held together by phosphodiester linkages from the 3’ of one nt to 5’ of another
> nt seqs written 5’ to 3’
> backbone = phosphates + pentoses / side groups = bases.
define the job of each of the three key RNAs
> mRNA = blueprint for AA seq
> tRNA = carry AAs + form long chain of AAs
> rRNA = part of ribosome → where peptides are made.
What are the three stages of the flow of genetic information?
> replication = DNA duplication and distribution evenly into daughter cells via cell division
> transcription = RNA synthesis
> translation = protein synthesis using mRNA, tRNA, and rRNA
what did Linus Pauling do in the race to define the structure of DNA?
Pauling devised 3D ball-and-stick models to deduce the structure of a folded protein - the alpha helix and beta sheet
what did Rosalind Franklin and Maurice Wilkins do in the race to define the structure of DNA?
> subjected crystallized DNA to x-ray beams
> spacing of atoms within crystal produces diffraction pattern
> diffraction pattern → mathematically interpret to structure elucidate
what did Edwin Chargaff do?
DNA base composition analysis for structure elucidation
how did Chargaff analyze base composition of DNA?
> extracted chromosomal DNA
> added protease to remove protein
> acid-treated to hydrolyze bases from strands
> subjected isolated bases to paper chromatography
> extract bands and determine amount by light spectroscopy
what is Chargaff’s rule?
A/G amount = T/C amount
what did James Watson and Francis Crick do?
> initially paralleled two backbones with like bases aligned
> didn’t fit the model
> ultimately matched bases as per Chargaff’s rule
who was the Nobel prize awarded to, and what would the Watson-Crick model be impossible without?
> Watson, Crick, and Wilkins
> Franklin’s x-ray diffraction data was key
describe each of the three models for DNA replication
conservative model
> one completely old w/ two parent strands + one completely new w/ two daughter strands (both identical)
+ implies re-association of parental strands post-replication
semi-conservative model
> each DNA has one continuous parental strand and one continuous daughter strand
dispersive model
> each DNA has pieces of parental and daughter strands
+ implies cleavage of parental strands during replication and subsequent reassembly
explain the 1958 Meselson and Stahl experiment
> used N because of the abundance of N in bases
> grew E. coli in media containing 15NH4Cl (ammonium chloride) as soul source of N
> cells generate NAs and nts with heavy nitrogen and not light nitrogen
> added these E. coli cells to media containing 14NH4Cl
> sampled cells at various time-points with DNA extraction
> the particles that has density similar to a specific region in the tube will be positioned around that specific region
> 14N DNA, whose density is light, will be around the region with light density
> 15N DNA, whose density is heavy, will be around the region with heavy density, allowing DNA replication model analysis
after one full replication, which replication model was eliminated, and why?
> photographed result showed one half-heavy, half-light band.
> indicates that both synthesized DNA molecules had half-light and half-heavy nitrogen
> disproves the conservative model, which states that one synthesized DNA molecule is full light and one synthesized DNA molecule is full heavy
after two full replications, which replication model was eliminated, and why?
> photographed result showed one band w/ light DNA and one band w/ half-heavy DNA
> indicated that two of the total DNAs had half-light and half-heavy
> indicated that other two of the total DNAs had full light
> disproves the dispersive model, which states that all four total DNA molecules would have ¼ 15N and ¾ 14N
what are the three steps of DNA replication?
> replisome binds + initiates synthesis
> rep forks continue synthesis in opposite directions
> rep forks hit terminus of replication and collide, releasing two chromosome copies
what did John Cairns do?
> fed E. coli with radioactively labeled thymine
> tracked its DNA rep via autoradiography (exposure of radioactive material to film)
> what does the bacterial chromosome look like?
> what is the size of DNA expressed as?
> what is the size of RNA expressed as?
> what does 1kbp equal to?
> what does 1Mbp equal to?
> what is the uncoiled length of E. coli’s genome in Mbp and mm?
> single molecule of circular dsDNA.
> size of DNA is expressed in base pairs.
> size of RNA is expressed in bases.
> 1,000 basepairs = 1 kilobase pair = 1 kbp/kb.
> 1,000,000 base pairs = 1 megabase pair = 1Mbp.
> genome of E. coli is 4.64 Mbp and is 1 mm in length uncoiled.
> what are the two levels of twisting in DNA? differentiate them.
> what assists in the second level of twisting in DNA?
> in the state of DNA once it has gone through the second level of twisting, do biological processes happen here or not?
> the first level of twisting is the DNA double helix twist that we normally see
> the second level of twisting involves additional twists (i.e. supercoils/supertwists) of the double helix (no base-pairing)
> DNA-binding proteins assists in supercoiling.
> in supercoiled state, there is not much biological processes → gene regulation
differentiate genotype and phenotype
> genotype = specific set of genes
> phenotype = set of all observable characteristics,, dependent on genotype + can be influenced by the environment
do prokaryotes need plasmids for their survival? why or why not?
> plasmids generally do not have genes for survival across all possible environments
> no
if prokaryotes don’t need plasmids for survival, what do plasmids have to offer for prokaryotes?
> virulence, antibiotic resistance, or special peptides that kill other bacteria for food (i.e. bacteriocins)
+ beneficial for their own good but also for the environment, such as nitrogen fixation and bioremediation properties
> what does the structure of plasmids look like in shape?
> how long are plasmids usually?
> are bacteria usually restricted to one specific type of plasmids and number of said type?
> most plasmids are circular, but many are linear
> 1 kbp to ≥ 1 Mbp
> some bacteria can contain different plasmid types + average number of plasmid copies present in the one cell
are the replication processes of plasmids and chromosomal DNA the same?
plasmids have their own replication functions compared to chromosomal DNA, but they still rely on host enzymes for replication