IMED1002 - DNA Variants 1 and 2 (L25 and L26)
1/43
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
44 Terms
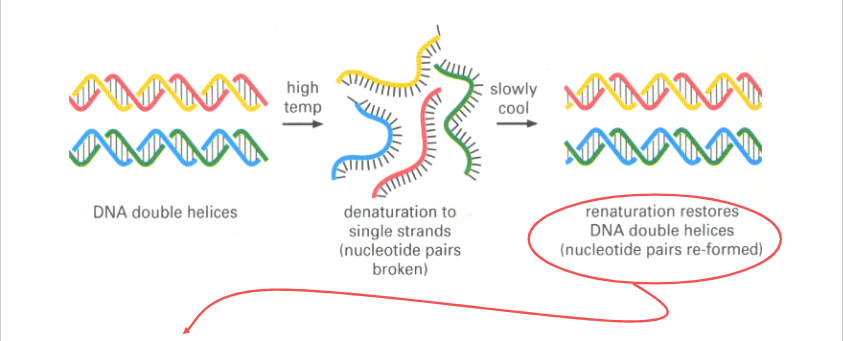
NA Denaturation and Renaturation
Hybridisation can occur between:
- Complementary DNA Strands -> DS DNA
- Complementary RNA Strands -> DS RNA
- Complementary DNA and RNA Strands -> RNA/DNA Strands
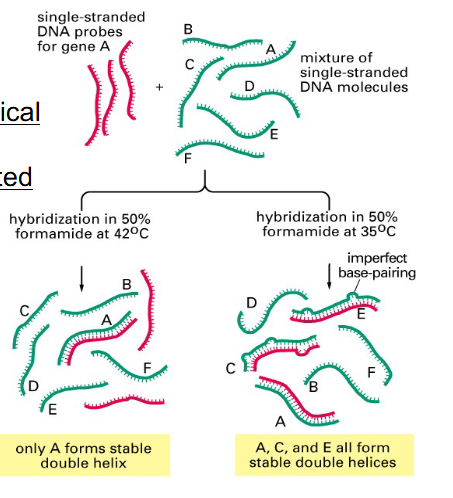
Nucleic Acid Hybridisation and Detection
Probes:
- 10s to 1000s nt long
- Homologous: detect identical nucleic acid molecules
- Heterologous: detect related molecules
Techniques:
- Southern blotting: when we have two DNA sequences binding to one another on gel and then blotting the onto a membrane
- Northern blotting: if we are looking for messenger RNA sequences, we separate out nucleic acids on gel and then transfer them, and blot them onto a membrane (to detect mRNAs)
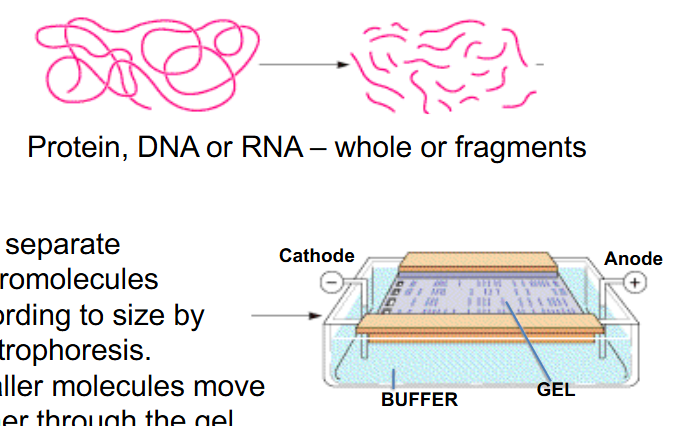
Gel Electrophoresis
- Protein, DNA or RNA - whole or fragments
- Can separate macromolecules according to size by electrophoresis.
- smaller molecules move further in the gel
- sodium dodecyl suplhate (SDS) is added

Transfer of Macromolecules from Gel to Membrane
- start with gel electrophoresis -> then blotting onto a membrane (easier to play with then gel)
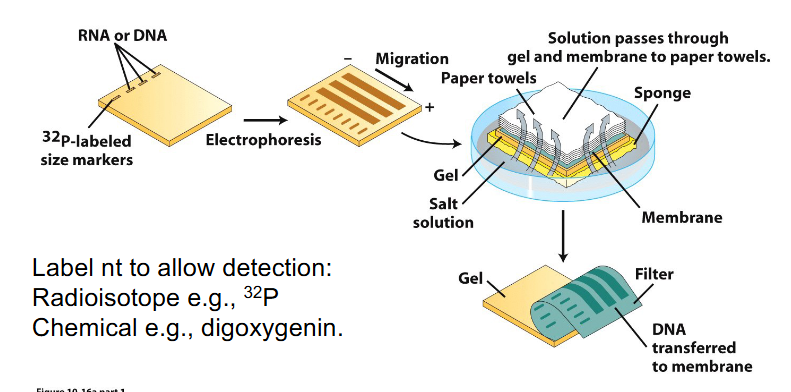
Southern and Northern Blot Analysis of NA
- method for detecting a molecule within a mixture
- Label nt to allow detection: Radioisotope e.g 32P or chemical e.g digoxygenin
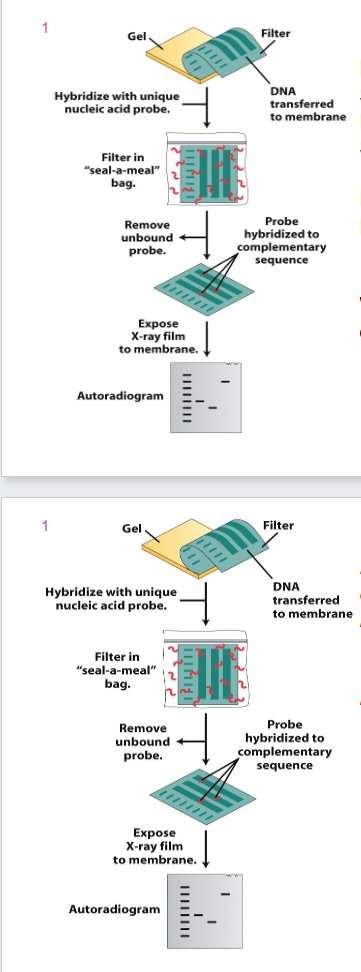
DIAGRAM OF BLOT ANALYSIS
- Procedure is termed Southern blotting when DNA is transferred (Edwin Southern)
- Northern blotting when RNA is transferred
- Western blotting is transfer of protein to a membrane
- irrespective of which tissue you get info from, you will get the same DNA (from heart, brain, muscle etc) with a southern blot
- however, northern blot runs RNA samples you will get different results depending on type of tissue. Because different cell types produce different array of proteins and hence mRNAs
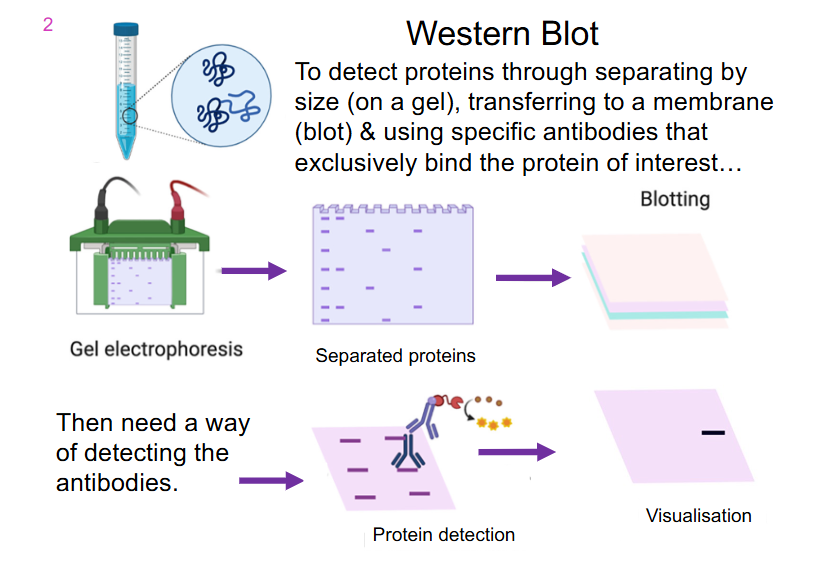
Western Blot
- to detect proteins through separating by size (on a gel), transferring to a membrane (blot) and using specific antibodies that exclusively bind the protein of interest
- in this example, we have an antibody binding to a protein which is then recognised by another antibody that allows us to form some sort of colour reaction
- western blot is literally the capacity to detect whether or not a specific protein is present in a sample
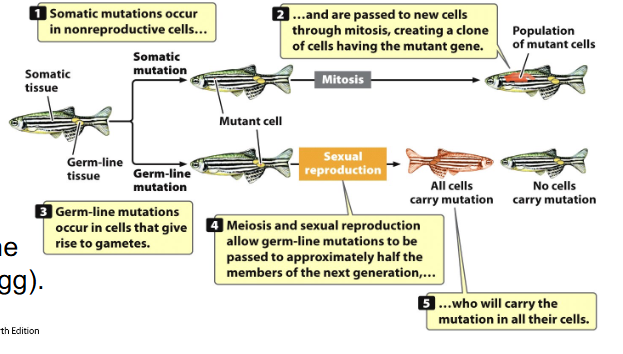
Mistakes can occur at many levels (Translation Errors, Transcription Errors)
- Translation Error: protein affected
- Transcription Error: mRNA and protein affected
- these only affect that cell - 'mutation' is not inherited (somatic)
- however, if there are errors in DNA replication - DNA, RNA and protein affected
- DNA variants can be passed to other cells and cause permanent change in a population of cells, and may be inherited if variant is in germline (sperm or egg)
Variants and Controlling Regions
- can occur anywhere in the DNA
- genes not only have coding regions but also controlling regions
- controling regions: direct DNA pol to initiate and terminate transcription, impact transcription factor binding, signal splice donor and acceptor sites, define intron/exon boundaries and splicing, direct 5' capping and 3' polyadenylation, affect ribosome binding
- changes in these regions can impact gene expression and function
DNA Variants can be beneficial
- May be beneficial: evolution
- May be deleterious (cause harm):
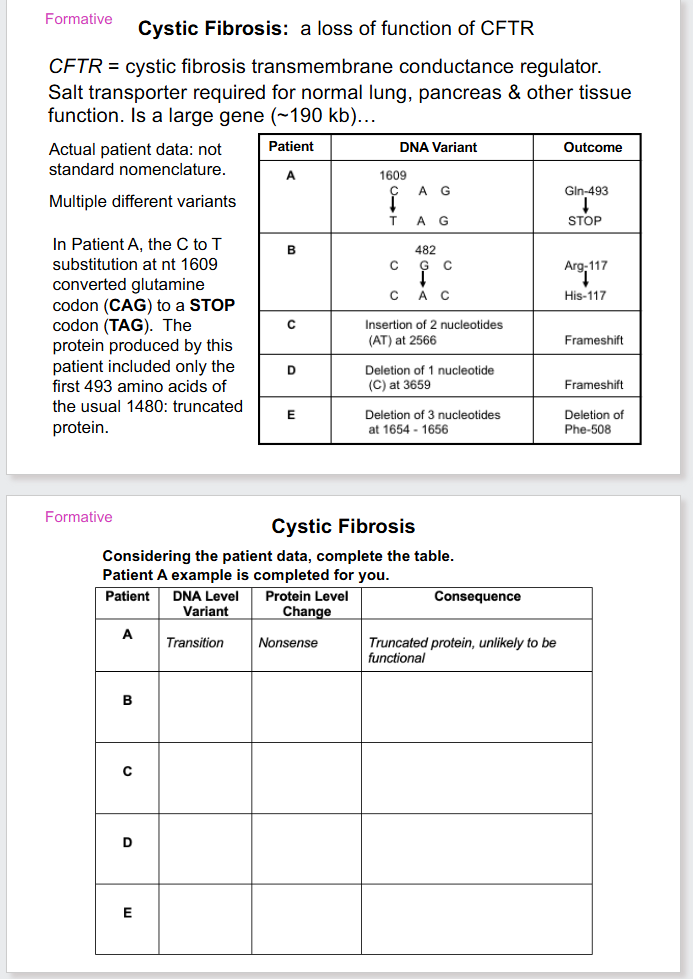
- loss of function variant - complete or partial absence of normal protein function (e.g cystic fibrosis)
- gain of function - entirely new characteristic or this appears in inappropriate tissue or time in development (e.g hyperactivity of a kinase in cancer)
- Lethal - causing premature death (e.g loss of a whole chromosome in early embryo)
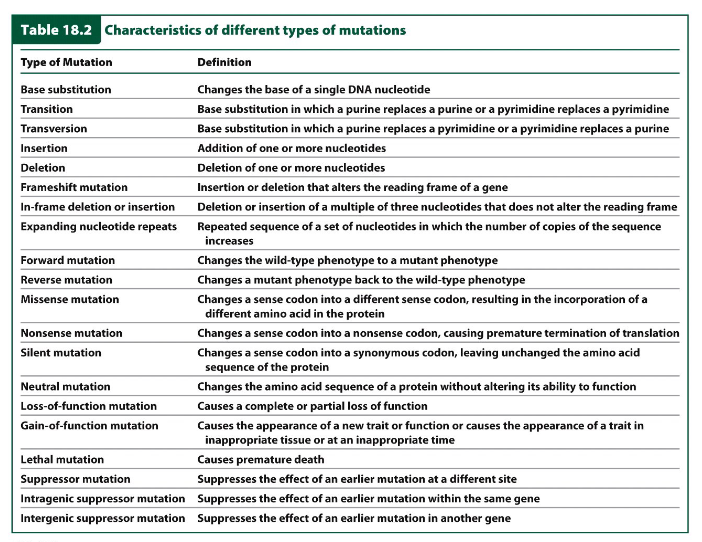
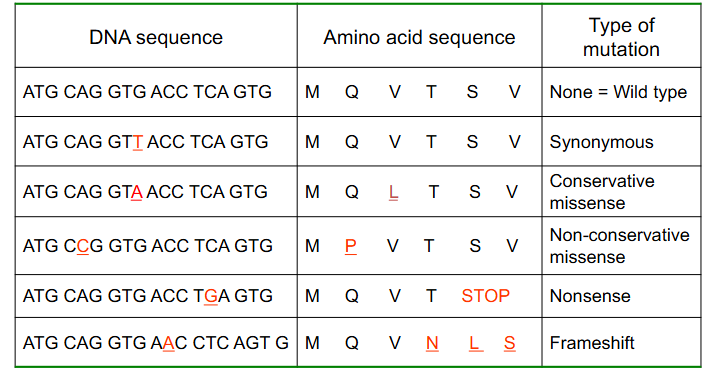
Characteristics of Different Types of Mutations
DIAGRAM ON SLIDE 16
Variations in DNA sequence (mutation)
- change in the genome - can occur spontaneously
- produce genetic variation (polymorphism)
- gene variation: change within a single gene. Some variants are point changes, in a single nt pair
- Alleles: alternate forms of a gene. 1 maternal, 1 paternal allele for genes on autosomes
Polymorphism
- natural variations in DNA that have no adverse effects on the individual and occur with fairly high frequency in the general population
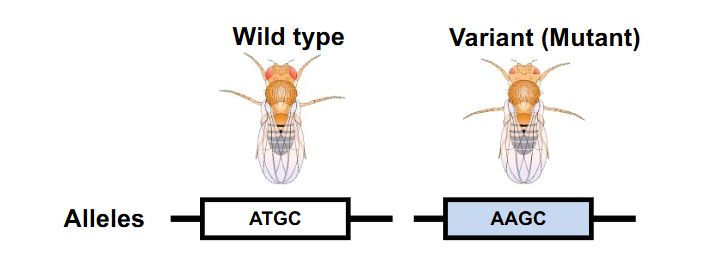
Wild Types vs Variants
- Wild Type: Standard/found in nature/laboratory stock
- Variant: Altered gene/s or characteristic/s (due to maturation)
- Phenotype (trait): the physical expression, appearance or manifestation.. "characteristic"
Variants at the DNA level classified by type
- Substitution: base is replaced by another base
- Deletion: one or more DNA base pairs lost
- Insertion: One or more DNA base pairs added
- Inversion: 180 degree rotation of piece of DNA
- Chromosomal rearrangements: can affect multiple genes
- Translocation: segment/s of chromosomes change places
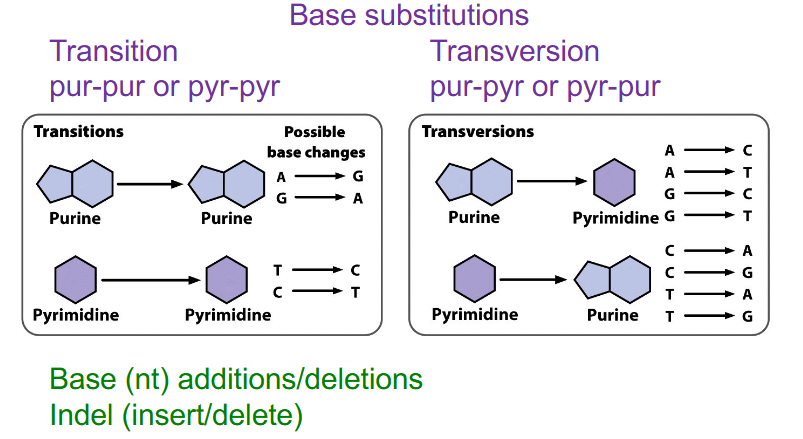
Two main types of point variants
Base substitutions
- Transition: pur-pur or pyr-pyr
- Transversion: pur-pyr or pyr-pur
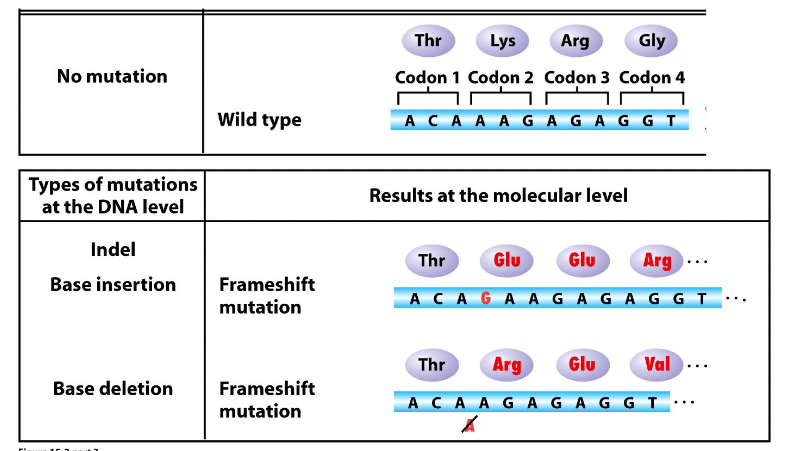
Indel
insertion or deletion
- base additions/deletions
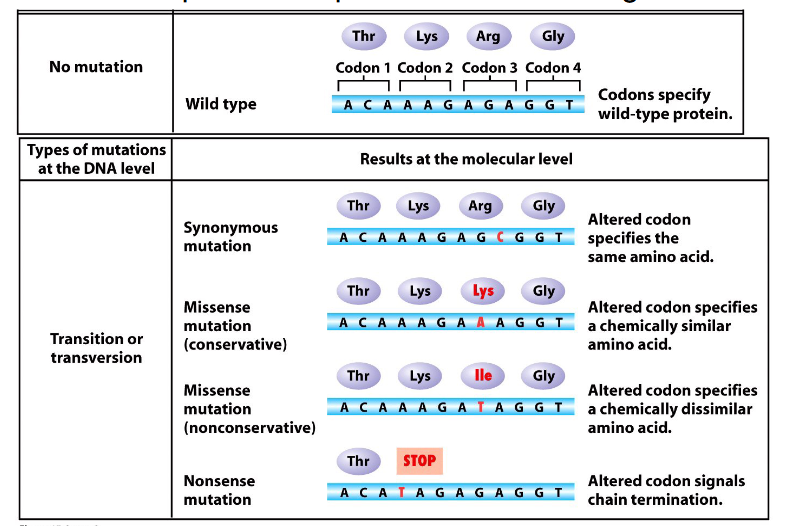
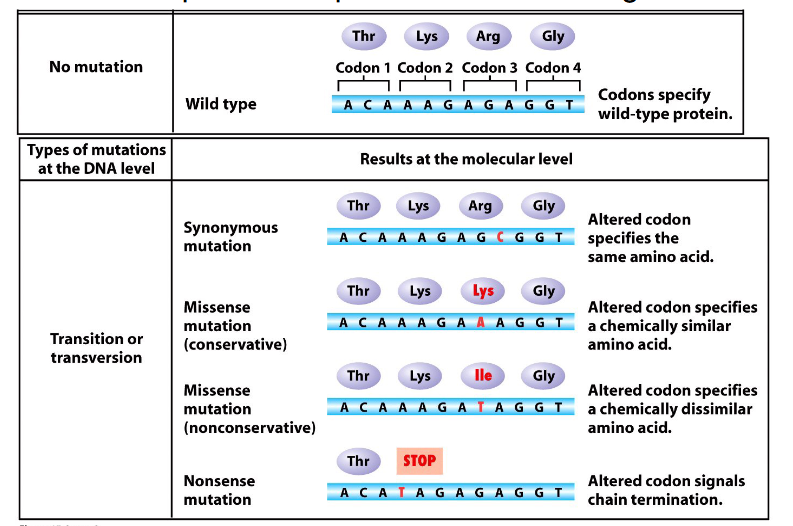
Consequences of Point Variants
- genetic code is redundant but not ambiguous
At the protein level variants:
- Synonymous (silent)
- Missense (conservative or nonconservative): different amino acid. Conservative missense means amino acid substitution is of a similar chemistry, nonconservative means its not
- Nonsense: insertion of a stop codon
- Frameshift: indel change, incorrect reading frame
- TAG or UAG is the stop codon
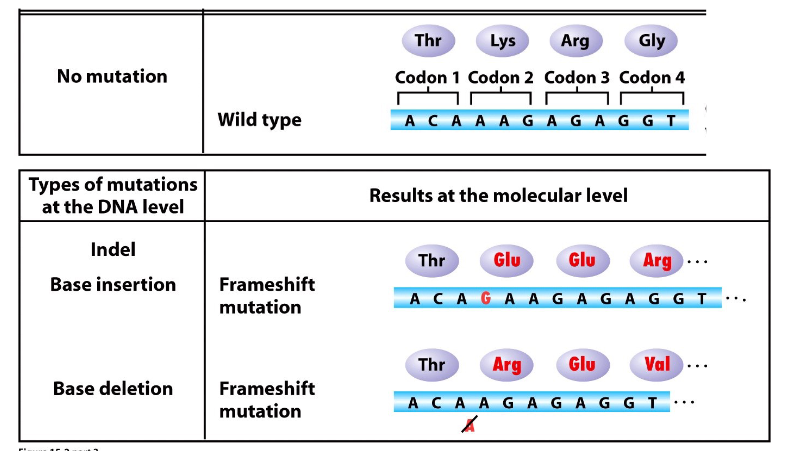
Indel Mutations
DIAGRAM ON SLIDE 23
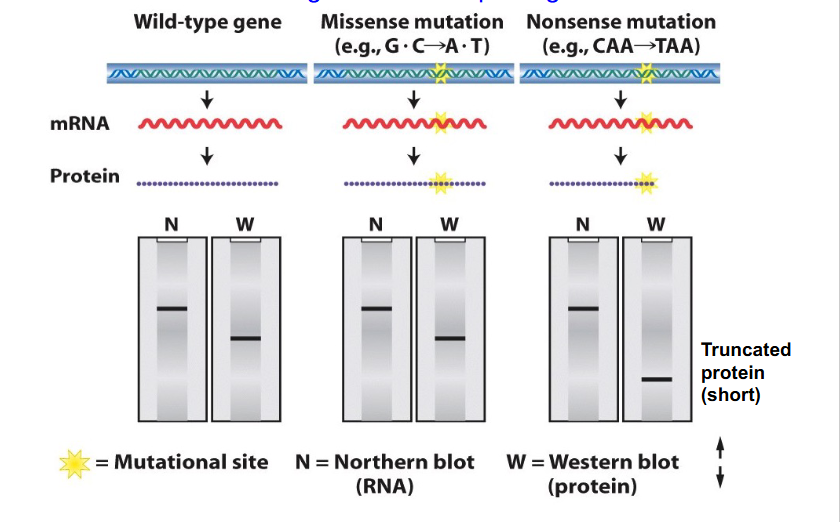
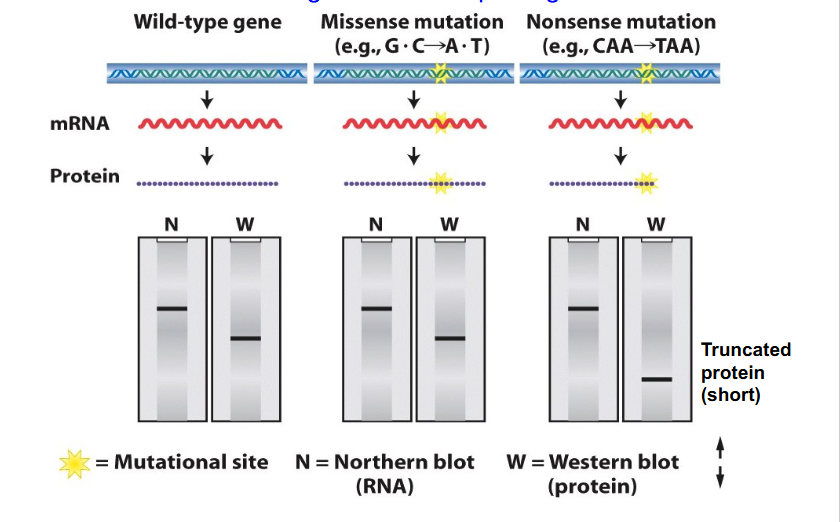
Blotting and Missense and Nonsense Mutations
- we can separate proteins, mRNAs and DNA according to their size on a gel and transfer proteins, DNA and mRNA onto a membrane then probe it with a particular sequence in the case of RNA that will hybridise to the corresponding complementary sequence or use an antibody to detect a particular protein
- if we have a missense mutation, this will change the sequence, which affects the mRNA and protein, but it will still give the same number of nucleotides and same number of amino acids and so visually based on blot diagram we would not be able to say if it was a missense mutation, whether or not it was wild type or the variant
- if we had a nonsense mutation, number of nucleotides would be the same so nucleotides in the message would be unchanged from the wild type. However, protein become shortened and hence run a further distance along our western blot
FINAL MESSAGE:
- A northern blot is not diagnostic of a point change within DNA or mRNA, but if there is a change to the length of protein due to stop codon insertion we can detect that on Western blot
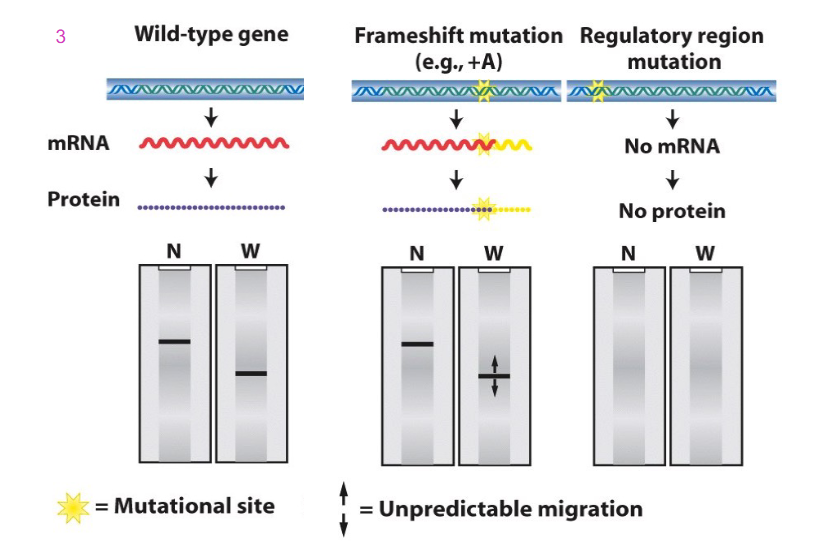
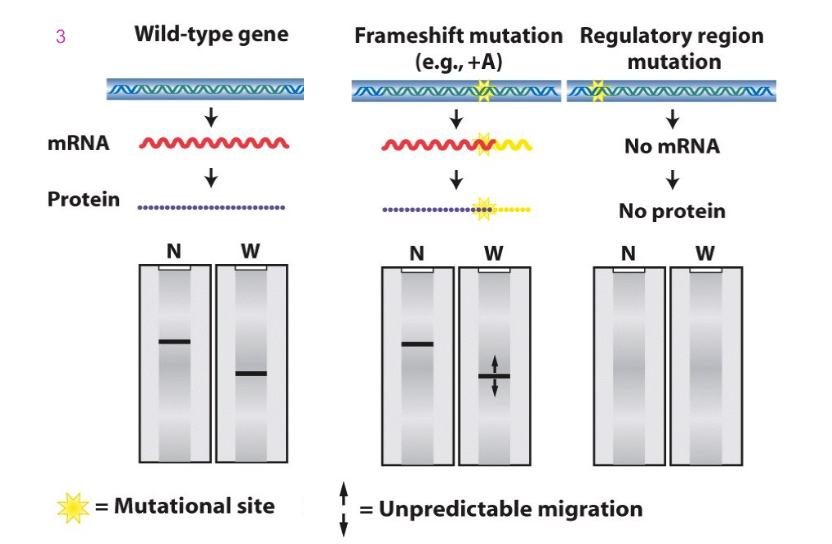
Blotting and Frameshift and Regulatory region mutations
- a frameshift mutation, e.g addition of base could cause a change to sequence of mRNA which might not be detected because the length of the message might not change. however, frameshift mutation can change the sequence of amino acids. And depending on whether or not that causes the stop codon to be recognised and termination to occur at the usual sign, we get the same length. it might cause a pre-mature stop codon, which would make nonsense and cause shorter protein to be made, or it might make longer protein
- if we have a mutation in the regulatory region (e.g TATA box) then we might not get assembly of general transcription factors and RNA polymerase 2. No transcription and hence no mRNA and hence no protein (no result on the Northern nor western blot)
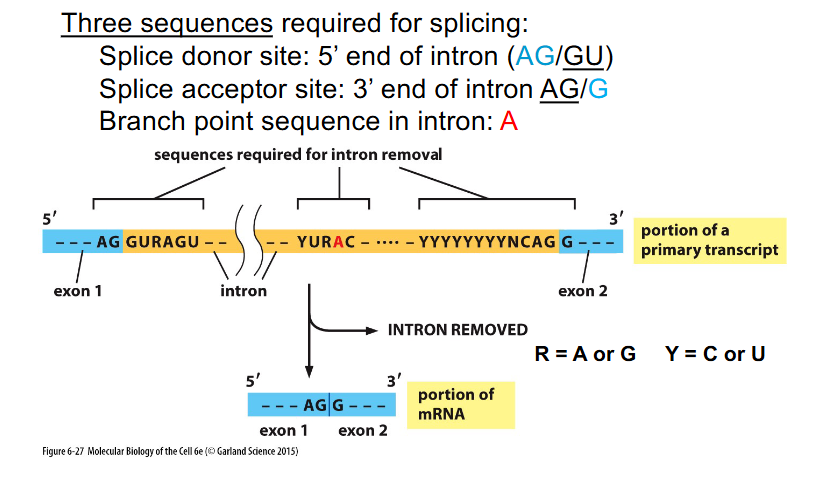
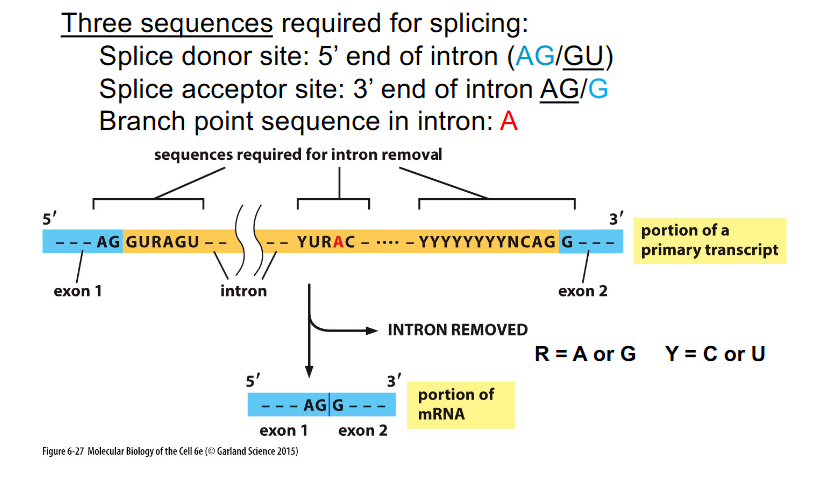
Sequences Required for Splicing
THREE SEQUENCES REQUIRED FOR SPLICING:
- Splice donor site: 5' end of intron (AG/GU)
- Splice acceptor site: 3' end of intron (AG/G)
- Branch point sequence in intron: A
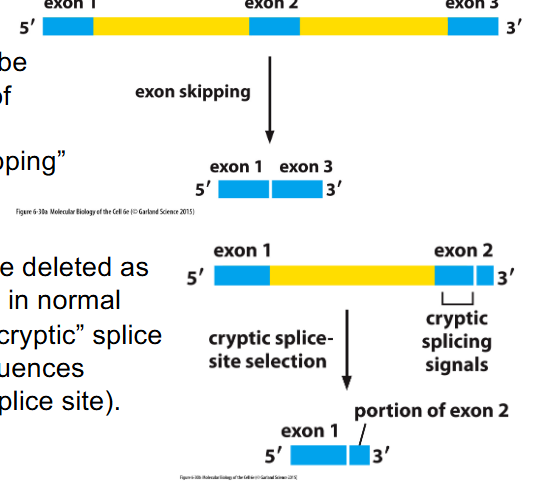
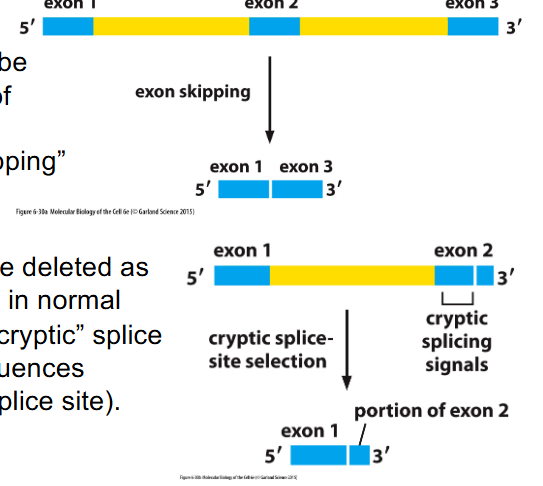
Splicing errors can be deleterious
- whole exons can be deleted as result of variants in splice signals: "exon skipping"
- Part of exon can be deleted as a result of variants in normal splice sites -> "cryptic" splice site selection (sequences resemble proper splice site)
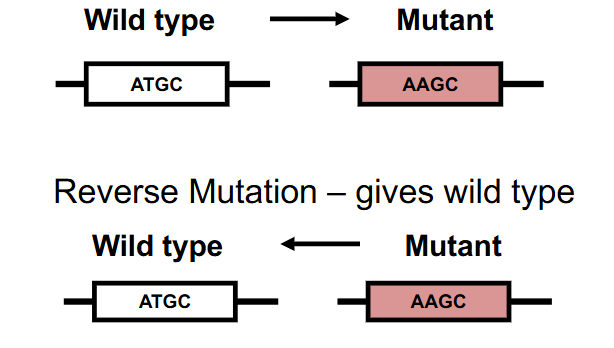
Forward Mutation
alters the wild-type phenotype to another phenotype
- Wild Type -> Mutant
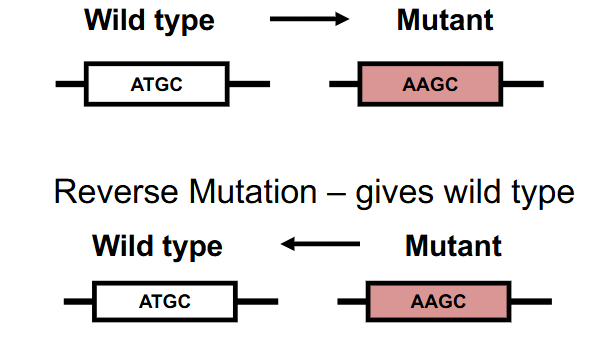
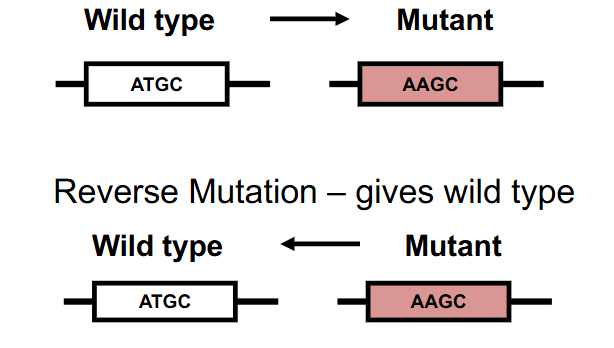
Reverse Mutation
changes a mutant phenotype back to the wild-type phenotype
- Mutant -> Wild Type
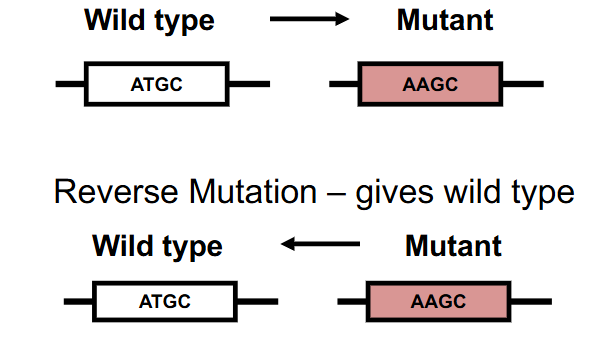
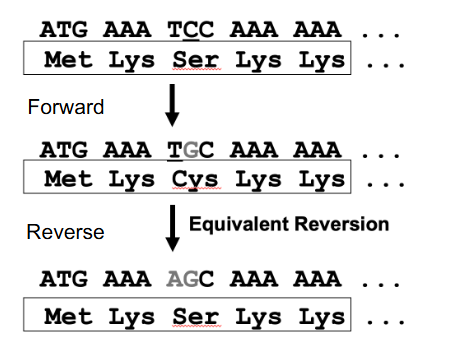
Reversion
- exact reversion (true or genotypic) gives the wild type DNA sequence
- Equivalent reversion gives a different DNA sequence, but wild type AA sequence (genetic code redundant): change in nt sequence to give wild type AA sequence, but not wild type nt sequence
- basically equivalent changes the forward mutation to something else that makes the same amino acid (genetic code is redundant)
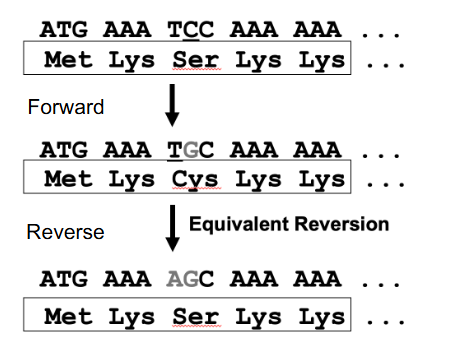
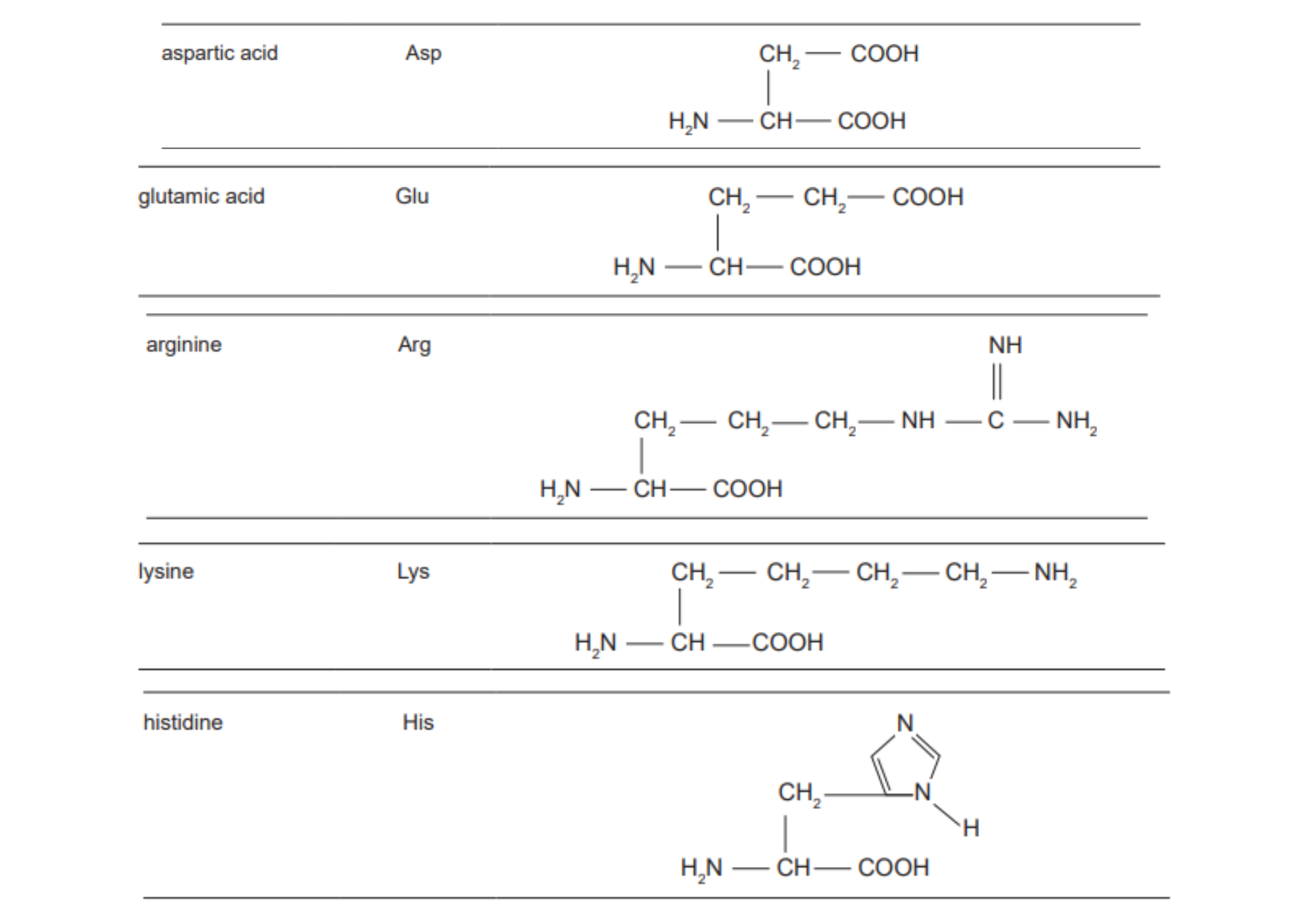
Acidic and Basic Amino acids
DIAGRAM ON SLIDE 31
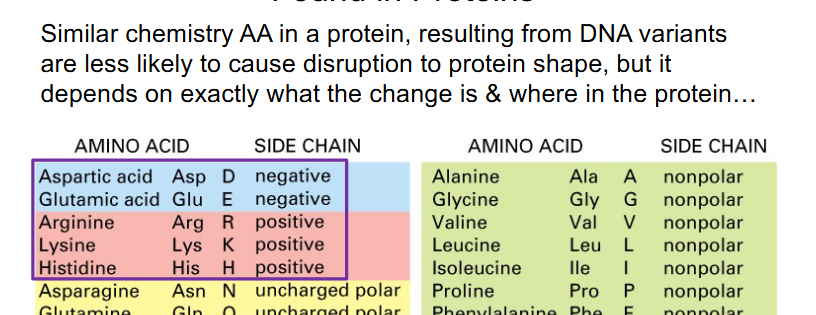
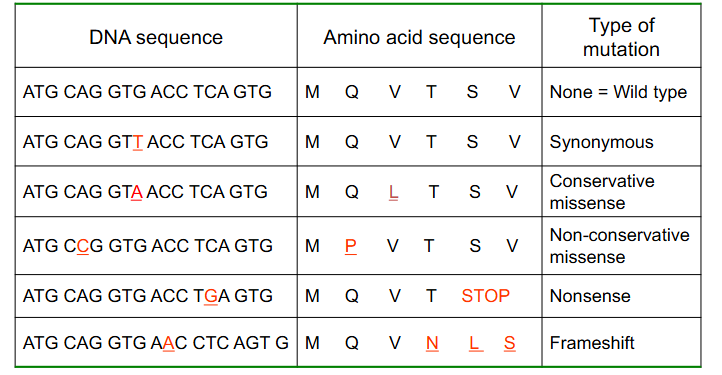
Point variations do not always have a phenotypic effect
- Other then synonymous, the least severe changes in a protein coding gene are missense, particularly conservative missense (conservative means amino acid replaced is similar structure and it doesnt have a major impact on the protein)
- however, some missense changes are deleterious
Nomenclature of Variants
WE KNOW THAT VARIANTS CAN ALTER ALL OF THE:
- DNA
- corresponding RNA
- translated protein's amino acid sequence
that means there is an enormous diversity of variants at nucleotide and amino acid sequence levels
- we will use the 3 letter abbreviations for amino acids (dont need to memorise)
Nomenclature
- the variants you need to know represent changes to the nucleotide base sequence on the DNA coding strand (c.) and the translated protein (p.) amino acid sequence
- We will designate the nucleotide number "base position" beginning with the first nucleotide of the translational start codon. i.e nucleotide #1 is the A in the ATG start codon on DNA coding strand. Hence nucleotide base 2 is T, and base 3 is G
- Bases 5' (more upstream) of the ATG start site are designated -1, -2, etc.
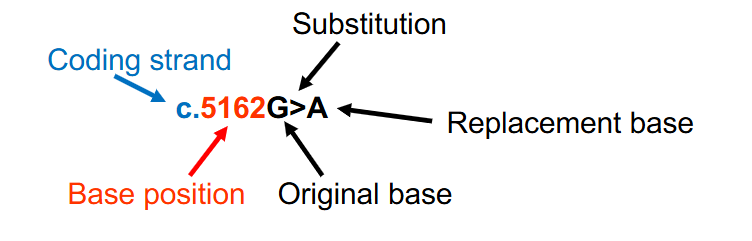
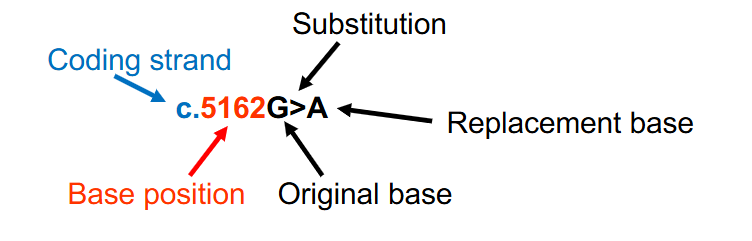
DNA Variant Nomenclature Example
- when there is a base substitution, both the original base and the replacement base are provided with an > between the bases to indicate the change
- c.5162G>A indicates that at nucleotide 5162 (with A of the ATG start codon being designated nucleotide #1) on the DNA coding strand, a guanine has been substituted for an adenine
- c. shows that its on the coding strand

DNA Variant Nomenclature for Deletions and Insertion
DELETION: c.197_198delAG
INSERTION: c.2552insT
- c.197_198delAG indicates that on the DNA coding strand, at nucleotide position 197 (with A of the ATG start codon being designated nucleotide #1) there is a two-base deletion of an A and a G
- c.2552insT indicates that on the DNA coding strand, at nucleotide position 2552 (with A of the ATG start codon being designated nucleotide #1) there is a one base insertion of T

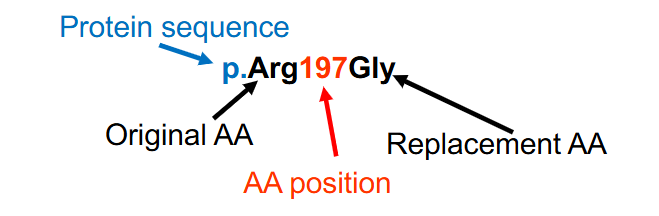
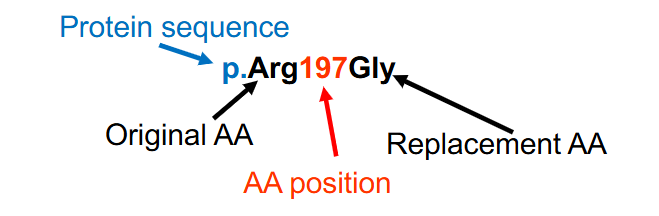
Protein Variant Nomenclature
- the protein's amino acids are numbered with the amino (N) terminal amino acid designated #1
- Both the original and the replacement amino acids are provided, using the 3 letter amino acid abbreviations
p.Arg197Gly (p.R197G) indicates a change from Arg (R, arginine) to Gly (G, glycine) at amino acid number 197
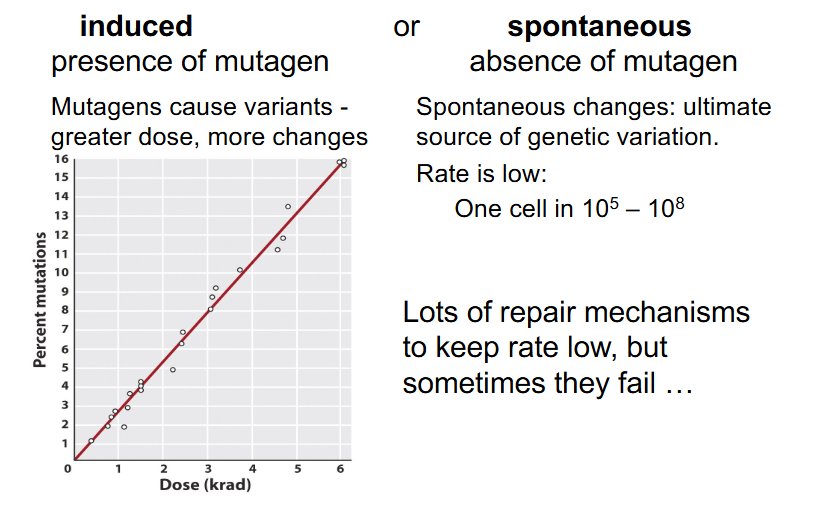
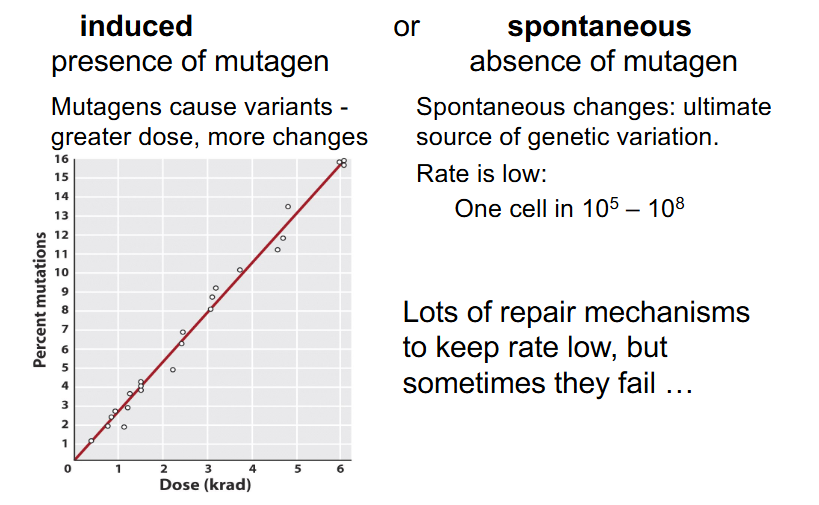
Origins of Variants/Mutations
- induced: presence of mutagen
- mutagens cause variants - greater dose = more changes
- spontaneous: absence of mutagen
- spontaneous changes: ultimate source of genetic variation
- Rate is low: One cell in 10^5 to 10^8
- lots of repair mechanisms to keep rate low, but sometimes they fail'
Mechanisms of Variant Induction
- mutagens can act through at least 3 different mechanisms:
- can replace a base in the DNA
- can alter a base and cause it to mispair
- can damage a base so it can't pair
- Additionally, mutagens can cause crosslinks between DNA molecules (between two separate DNA double helices), cause breaks in DNA
- mutagens have specifc modes of action and often cause a specific type of DNA change
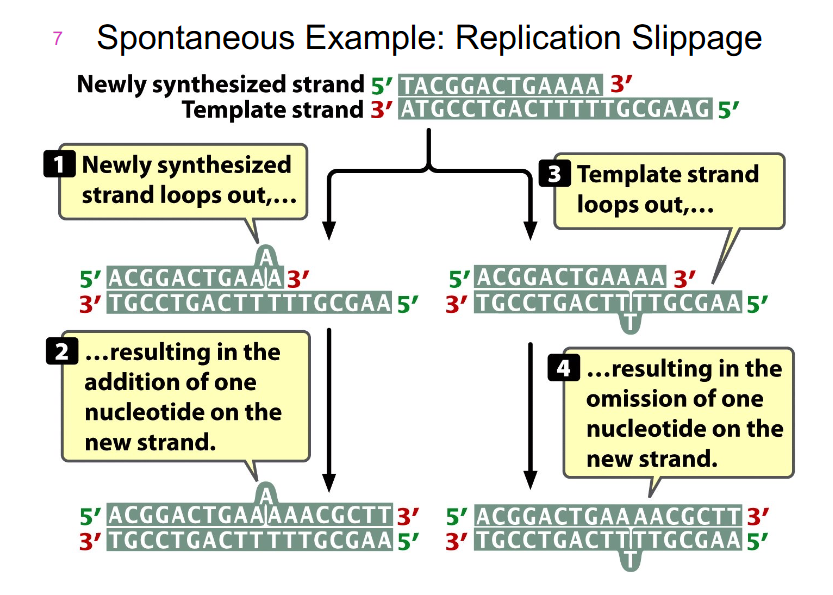
Spontaneous Example: Replication Slippage
- happens when there are repeated sequences along our DNA
- there are some ways we can slip during replication
- the newly synthesised strand can loop out, resulting in addition of one nucleotide on one new strand
- the opposite can happen if we have a slippage of template strand, resulting in the omission of one nucleotide on the new strand
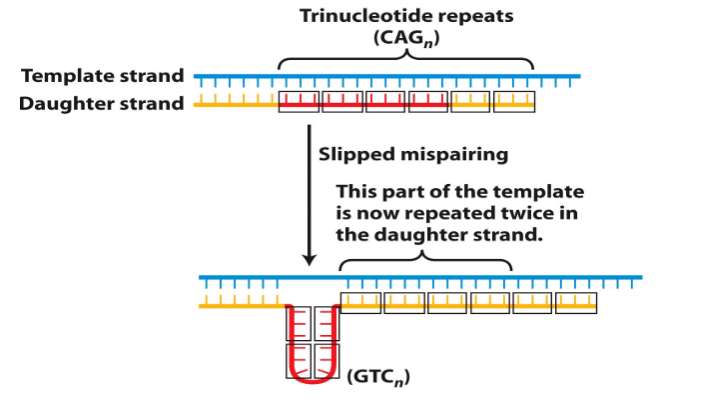
Trinucleotide Repeat Disorders
- common mechanism responsible for a number of genetic diseases is expansion of a 3bp repeat - hence the name
Slippage Consequences
NEW STRAND:
- when new strand slips it has an extra base that it will copy form original strand, which means that compared to the wild type strand it will have an added base (addition)
TEMPLATE STRAND SLIPPAGE:
- when template strand slips the new strand has one less base. This means that compared to the new strand, the new strand will have one less base (deletion)
Huntington's
- midlife onset of dementia, followed by death
- Caused by expansion of CAG (Codes for glutamine) repeats in the coding region of the Huntingtin gene
- CAG repeats translated into polyglutamine tract
- 6-35 repeats: no issue
- 36-121 repeats: Huntington's
- more then 70 repeats: Huntington's with earlier (juvenile) onset
- Other disorders (e.g spinocerebellar ataxia) also caused by CAG expansion in coding region
- Polyglutamine tract believed to cause neurotoxic aggregates (PolyQ diseases)
General Statements on DNA Repair Mechanisms
- Most important repair occurs during DNA replication: 3' to 5' exonuclease (proofreading) reduces DNA mispairing
- Are several complex pathways for DNA repair
- Most repair requires 2 DNA strands so that one can act as the template for synthesis of the other
- Redundancy, i.e most DNA damage can be repaired by more then one repair pathway
- ensures low level of DNA variation -> if one pathway fails to recognise and repair damage, another pathway may still act
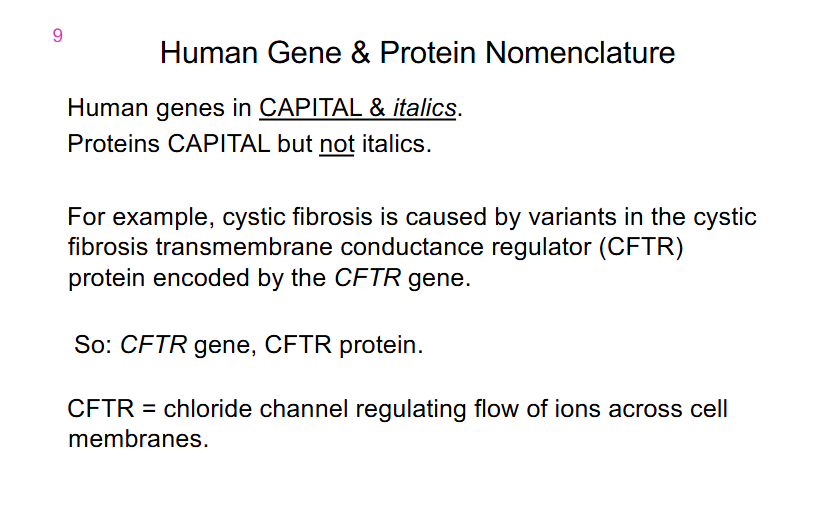
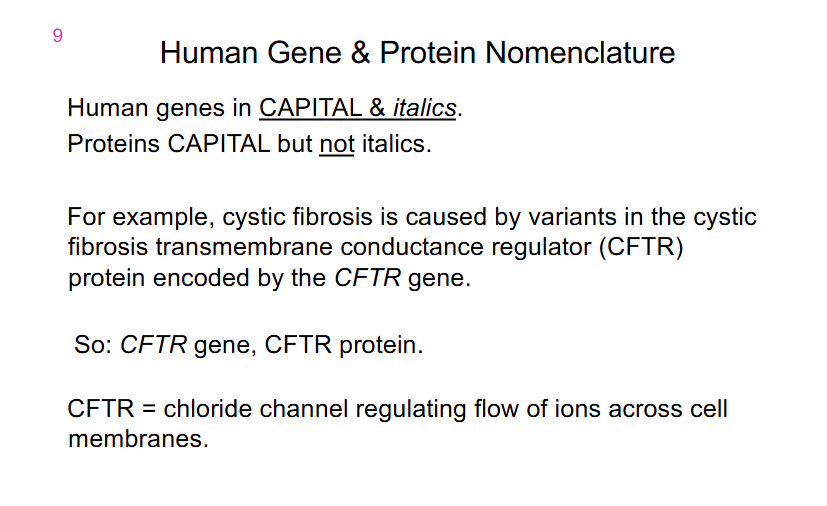
Human Gene and Protein Nomenclature
- Human Genes in CAPITAL and italics
- Proteins CAPITAL but not italics
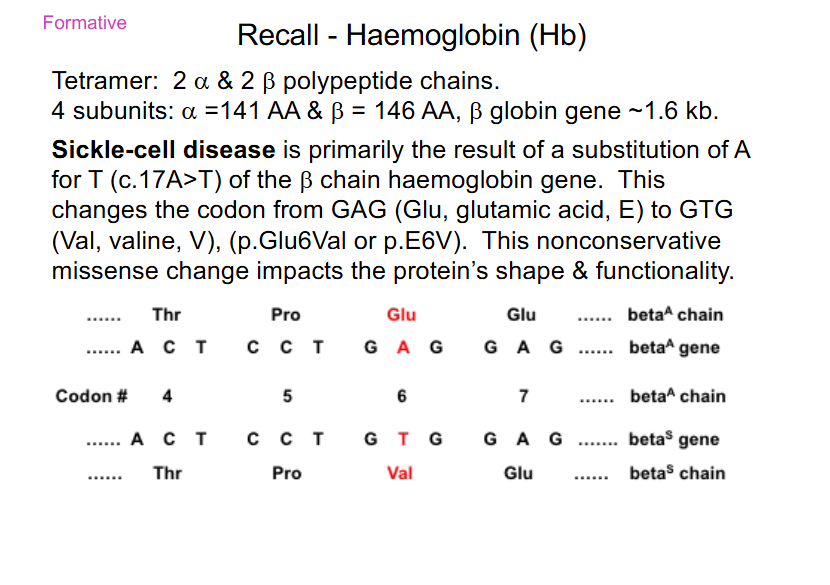
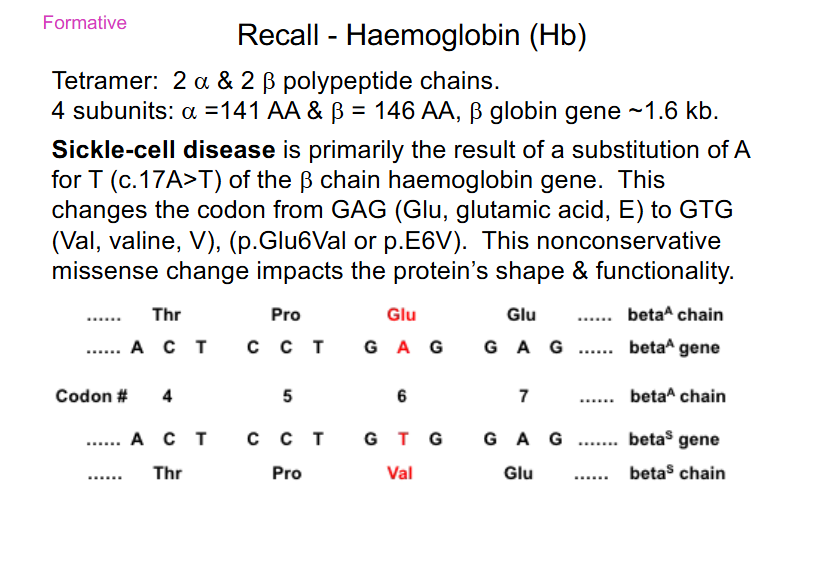
Sickle Cell Disease is caused by
- substiution of A for T (c.17A>T) of the beta chain haemoglobin gene
- Glu -> Val
- nonconservative missense: AA change from acidic, polar (Glu) to a Val (neutral and non-polar)
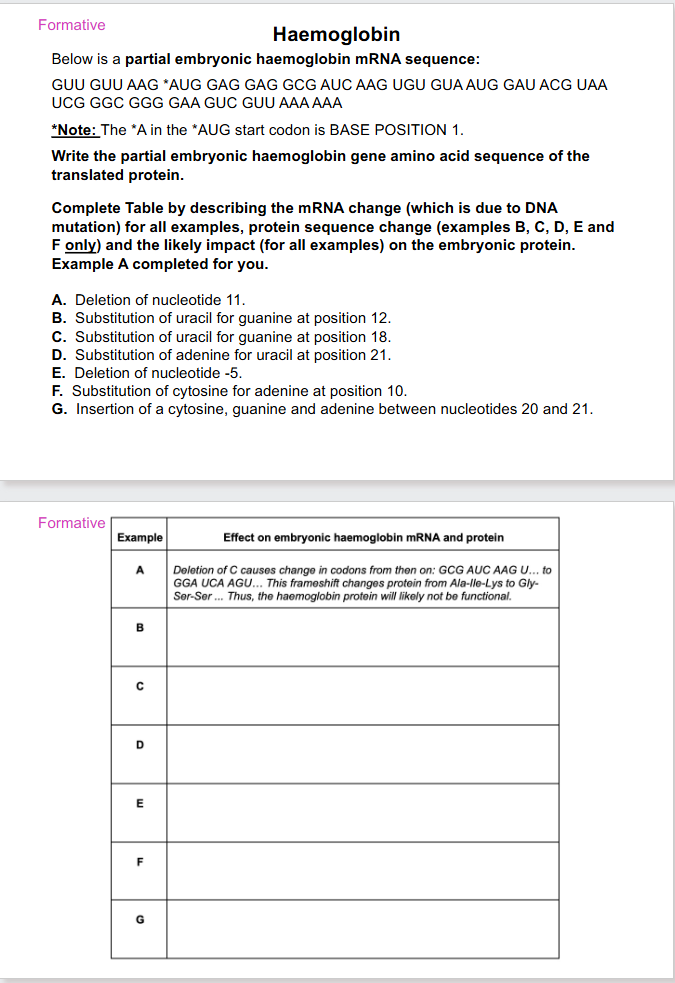
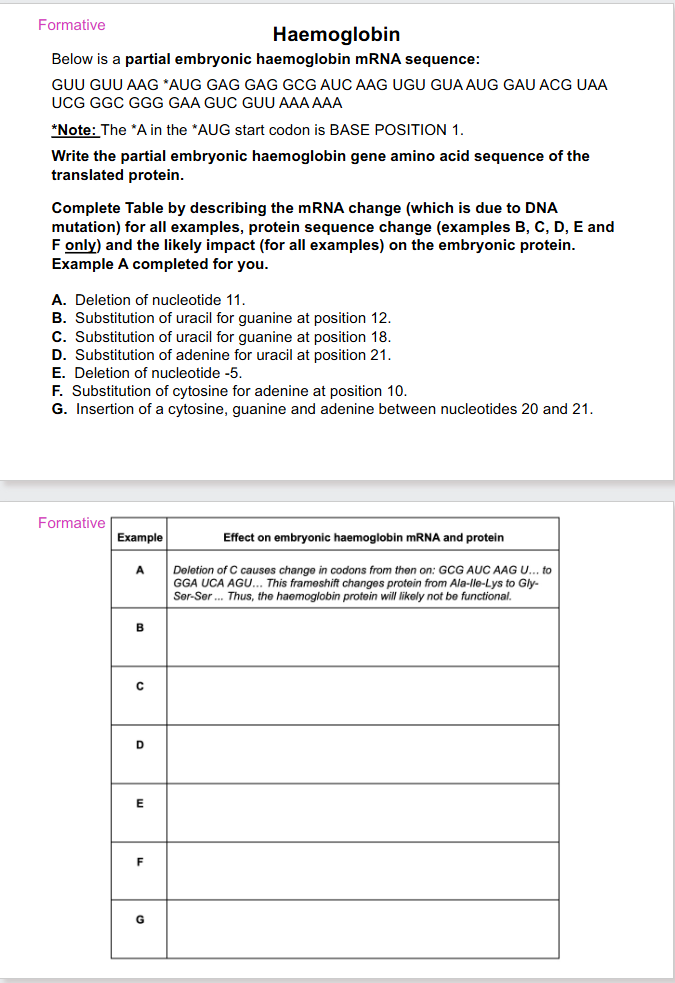
SLIDES 52 TO 55 ARE FORMATIVE
DO THEM
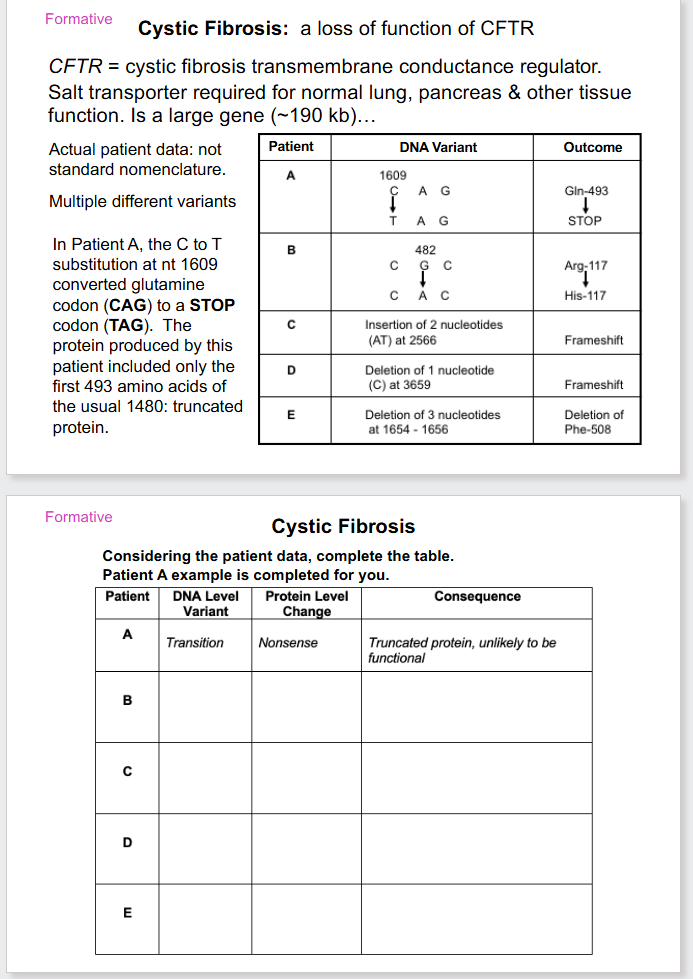
CS QUESTION
DO THEM