MYCO 2.1A,B Techniques in Fungal Systematics, Kingdom Mycota
1/125
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
126 Terms

_ is an approach to species classification that combines multiple lines of evidence to delimit and identify organisms more accurately, which means that instead of relying solely on traditional morphological characteristics, this will incorporate diverse data sources, including molecular data, ecological behavior, and physiology.
Integrative taxonomy
What are the 2 most commonly utilized repositories for information on fungi?
Mycobank
Index Fungorum
Current estimates for fungal species = _
Currently described + culturable = _
Current estimates = 2.2 - 3.8 million or more species
Currently described + culturable = 146,000
Enumerate morphological assessment techniques for fungal identification
cdet
Culture (3 media, 3 temperatures)
Description (filamentous/colony, microscopic characters)
Extrolites profiling
Taxonomic keys
Cite 2 reasons that make fungal systematics complex
Many fungi have been described but do not have a cultured representative.
There are also many fungi submitted as new species, but these may have already been identified previously whether in their sexual/asexual forms, thus there is a need to check
T/F: There is no need for a renewed effort to develop fungal systematics
FALSE
There is a need to develop fungal diversity estimates through systematics
_ is the current method being used to estimate fungal diversity through systematics
Integrative taxonomy (morphology + molecular data)

Explain why there is a need to develop fungal diversity estimates through systematics
Current estimates = 2.2 - 3.8 million or more species
Currently described + cultured = 146,000
Fungal abundance cannot be overlooked for any region of Earth. Hence, there is a need for a renewed effort to develop fungal systematics.
One way we’re doing it now is through integrative taxonomy, combining morphological + molecular data
_ refers to the ratio of the vapor pressure of water in a substance to the vapor pressure of pure water at the same temperature, basically referring to how much water is “free” or available for biological and chemical reactions
Water activity
_ refers to organisms that thrive in environments with low water activity, often possessing adaptations like accumulating solutes (e.g., trehalose, glycerol) to retain water
Xerophiles
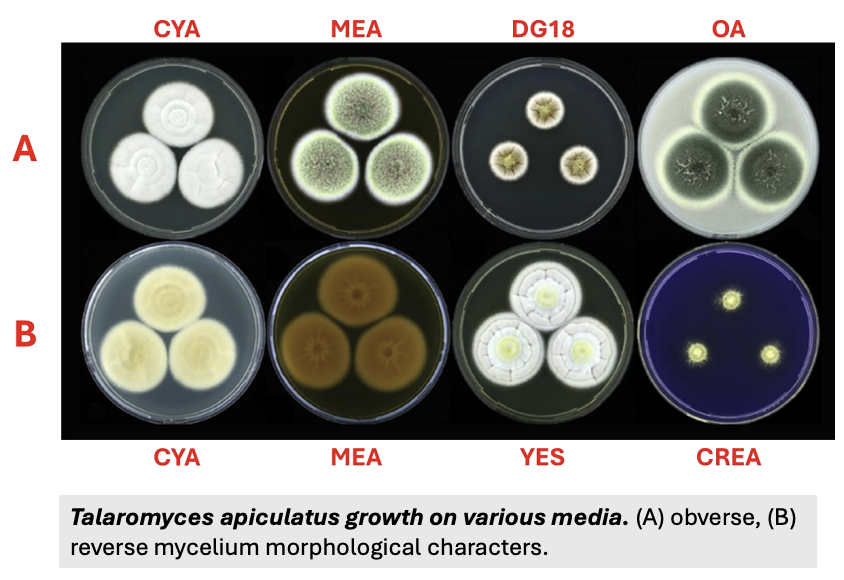
Fungi for identification are cultured in _
3 different types of media and temperatures
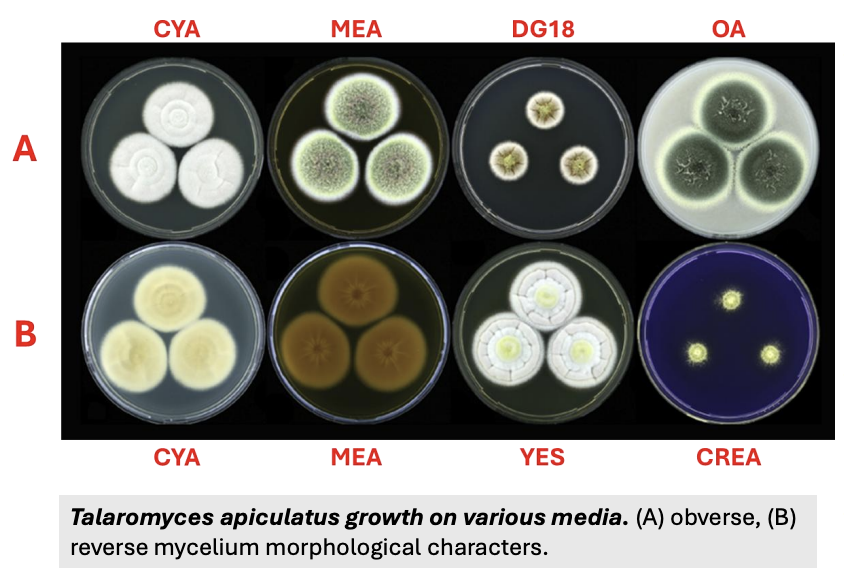
Explain the 3 different types of media and temperatures used for fungal identification (morphological assessment)
Media
CYA (Czapek Yeast Autolysate) agar = ic ff isolation and cultivation of heat-resistant filamentous fungi
MEA (Malt Extract Agar) = die ym detection, isolation, enumeration of yeast and molds from clinical and non-clinical samples
G25N (25% Glycerol Nitrate Agar) = selective growth medium for xerophilic fungi, which grow at low water activity or availability
Temperature (can be used for growth rate)
5 C = cold-tolerant
25 C = mesophilic (closer to 30 C in Asia)
37 C = thermotolerant (should not be increased to 40; would trigger death or microcycle sporulation)
Other media may be used depending on the goal of study, fungal group, etc.
Important to use at least 3 media, 1 mesophilic
Total = 9 setups (3 media, each with 3 temps)
T/F: Malt extract agar (MEA) and Potato dextrose agar (PDA) are generally a mesophilic type of growth medium for yeast and molds
TRUE
Why are 3 different media and temperatures used for fungal morphological assessment?
It’s bc the combination of their growth patterns, growth rates, and sizes of a fungus on these different types of media and temperatures can be noted down to be used as reference values for those only using morphological characters as their basis of delineation
Type of media for fungal morphological assessment
Selective growth medium for xerophilic fungi, which grow at low water activity or availability
G25N (25% Glycerol Nitrate Agar)
Type of media for fungal morphological assessment
For isolation and cultivation of heat-resistant filamentous fungi
CYA (Czapek Yeast Autolysate) agar
Type of media for fungal morphological assessment
For detection, isolation, and enumeration of yeast and molds from clinical and non-clinical samples
MEA (Malt Extract Agar)
Enumerate 3 temperature settings to which fungi are subjected to for morphological assessment
5 C = cold-tolerant
25 C = mesophilic
37 C = thermotolerant
Fungal description requires both _ characters
filamentous or colony and microscopic

Fungal growth diameters should be measured around _ days
7 to 14
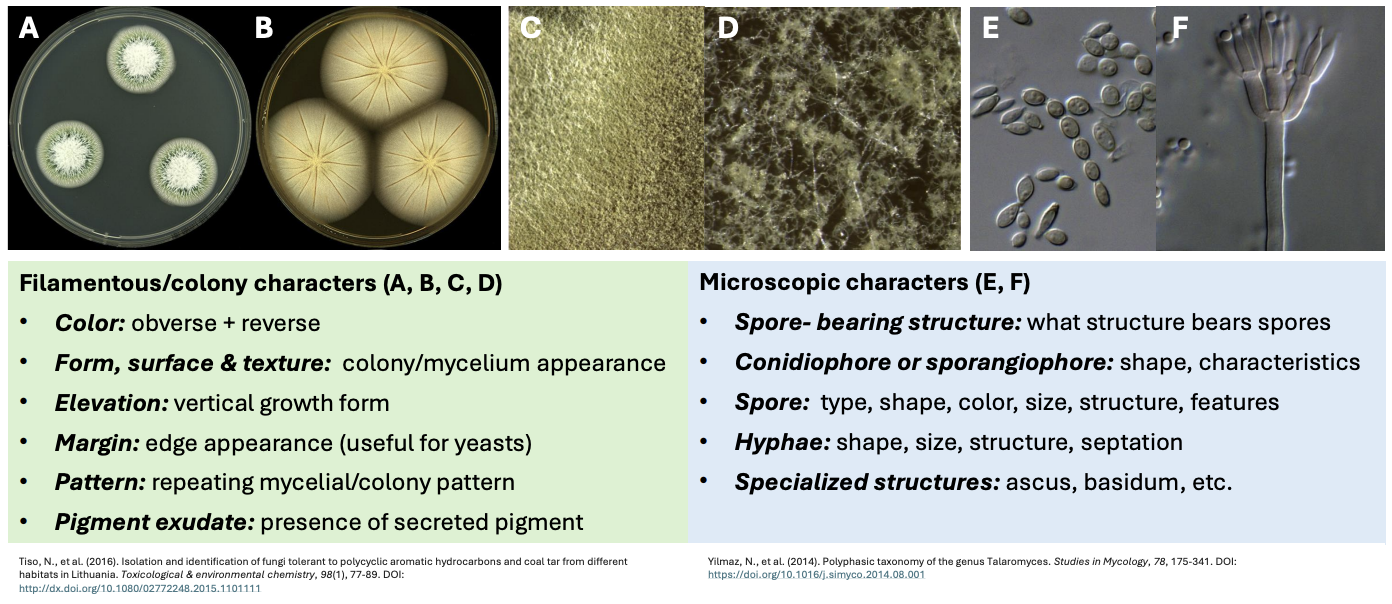
Explain filamentous / colony characters for fungal description
cfem 2p
Color = obverse (front of plate) + reverse (bottom)
Obverse = mycelial features
Reverse = secretory features
Form, surface, & texture = colony / mycelium appearance
Elevation = vertical growth form
Margin = edge appearance (useful for yeasts)
Pattern = repeating colony / mycelial pattern
Pigment exudates = presence of secreted pigments
To inoculate plates for fungal description, there should be _ (number) inoculation points, equidistant from the edges of the plate and from one another to make sure that when you inoculate from a single source, you’d be able to generate a pure culture
3
_ refers to any substance created within the cell and then transported outside
Extrolite

If you don’t know how to describe (do fungal descriptions), it is better to use _
taxonomic identification keys
_ are fundamental taxonomic criteria in filamentous and yeast fungi because the production of identified _ can be compared with known or published standards
Extrolite profiles

Explain microscopic characters for fungal description
scs hs
Spore-bearing structure = what structure bears the spores
Conidiophore or sporangiophore = shape, characteristics
Spores = type, shape, color, size, structure, features tsc ssf
Hyphae = shape, size, structure, septation shasi strucsep
Specialized structures = ascus, basidia, etc.

_ use morphological features to identify fungi and classify them
Taxonomic keys

_ is one of the most utilized taxonomic identification keys for lab-cultured fungi
_ is preferred for identifying mushrooms
_ is where you can access fungal taxonomic keys
Pitt & Hocking Keys
British Mycological Society Keys
Mycobank

Why is morphological taxonomic evaluation limited in fungi?
Convergent morphology: Many unrelated fungi exhibit similar macroscopic and microscopic features, making it difficult to distinguish species based solely on morphology
Phenotypic plasticity: Environmental conditions (e.g., temperature, substrate, and humidity) can influence fungal morphology, leading to variability even within the same species
Incomplete or missing sexual stages: Many fungi, especially those historically classified as Deuteromycota (fungi imperfecti), lack known sexual stages (teleomorphs), which were crucial for traditional classification
T/F: In invertebrates, morphology is the gold standard for species delimitation
TRUE
Why should molecular and morphological data go hand-in-hand for fungal species delimitation?
It’s bc the accuracy of molecular data is heavily dependent on the marker used, thus morphological traits must also be considered

T/F: A positive result in the ELISA setup shown in the image confirms that the antigen has two distinct epitopes recognized by different antibodies
TRUE
In sandwich ELISA, 2 different antibodies, one for capture and one for detection, must recognize different epitopes on the same antigen. Otherwise, if they recognize the same epitope, they would compete for the same binding site and thus prevent proper detection.
Explain ELISA as method for molecular identification of fungi
*Sandwich ELISA
Wells are pre-coated with capture antibody, then samples with fungal antigens are added
Capture antibody binds the antigen with high specificity
Primary detection antibody binds to the immobilized antigen (on a different site)
Enzyme-conjugated secondary antibody binds to Fc region of primary antibody, such that (for signal amplification)
Upon addition of substrate, enzyme catalyzes the substrate, causing a detectable color change in the presence of the specific fungal antigen
“Sandwiching” ensures high specificity, reducing false positive results, because 2 different antibodies must recognize the same antigen

Enumerate molecular techniques for fungal identification
ELISA
PCR
MALDI-TOF MS
*2 most common technique = ELISA, PCR

_ is an immunological method that can be used to target cell wall proteins and membrane proteins specific to different species of fungi
Enzyme-linked immunosorbent assay (ELISA)
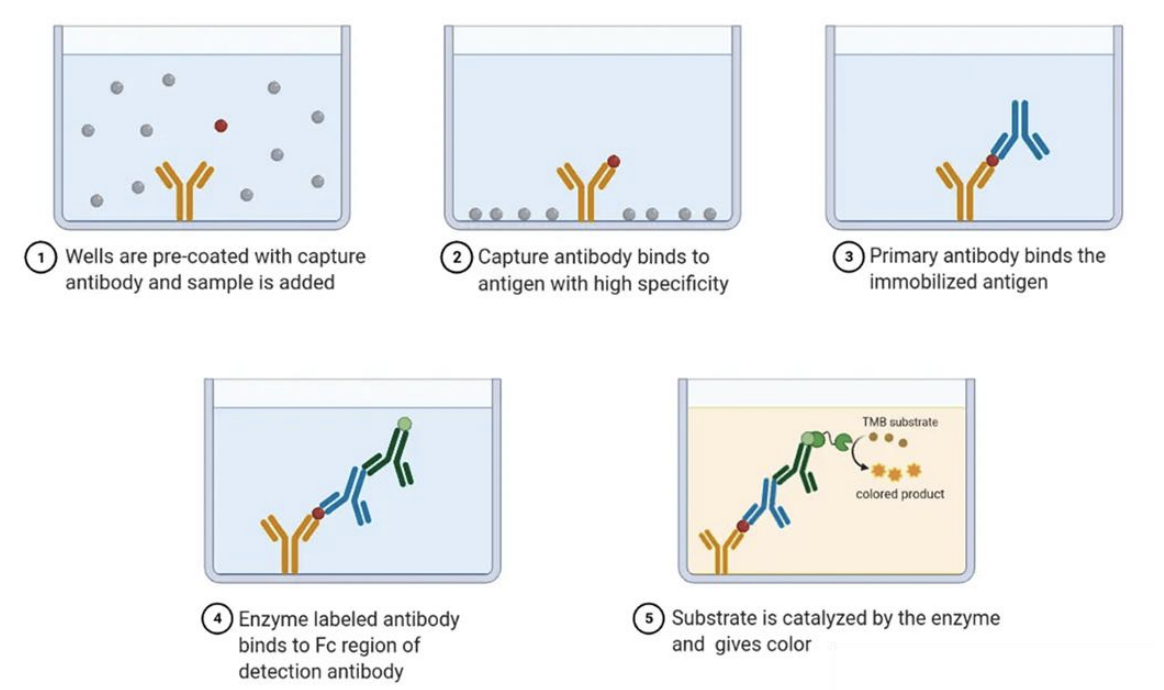
ELISA application
A research lab is developing a sandwich ELISA to detect a fungal pathogen in patient samples. However, during assay validation, they observed that even in fungus-free samples, some wells still show a color change after substrate addition.
As the lead scientist, you suspect an issue with antibody specificity. Based on your understanding of sandwich ELISA, propose two possible explanations for this false positive result and suggest a way to confirm your hypothesis experimentally.
Possible explanations
Nonspecific binding of the enzyme-conjugated secondary antibody i.e. may be binding directly to the capture antibody instead of the primary antibody, leading to a false positive signal.
Contamination of fungal antigens or cross-reactivity, i.e., detection antibody may be binding to a structurally similar antigen from another organism present in the sample
Experimental confirmation
Perform a control experiment where the antigen is omitted. If color still develops, the issue is likely due to the antibodies' non-specific binding of the antibodies.
_ is a rapid and low-cost method for generating molecular markers for sequence analysis and comparison in fungal identification
Polymerase Chain Reaction (PCR)
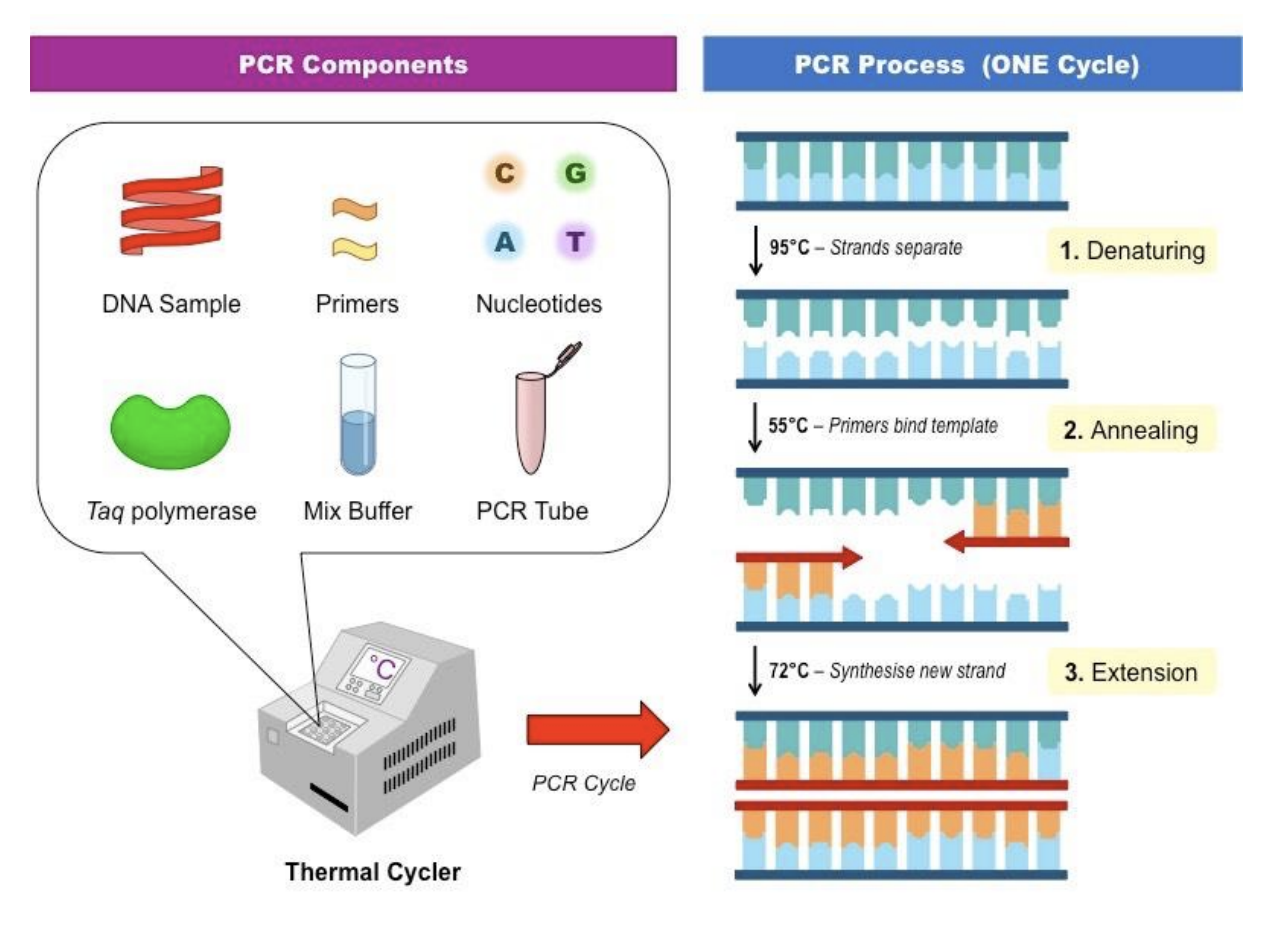
T/F: In terms of fungal identification, PCR is more specific in amplifying DNA sequences and targets compared to ELISA
TRUE
Explain PCR as method for molecular identification of fungi
PCR components dnp tmp
DNA template (Fungal DNA sample)
Nucleotides = A, T, C, G
Primers = short sequences that target specific genes
Taq polymerase = enzyme that synthesizes DNA
Mix buffer = ensures optimal reaction conditions
PCR tube = container
PCR process dae
Denaturation (95 C) = unwinding of DNA
Annealing (55 C) = primers binding to DNA template
Extension (72 C) = Taq polymerase synthesize DNA by adding nucleotides to primers
T/F: The DNA sequence of the forward primer is identical to the sequence of the final PCR product at the 5' end, while the reverse primer is identical to the product's 3' end
TRUE
T/F: The primer sequence is removed and degraded during each PCR cycle, so it is not present in the final amplicon
FALSE
The primer is actually incorporated into the final PCR product during extension and remains in the amplicon
T/F: Extrolite profiles are fundamental taxonomic criteria only in filamentous fungi
FALSE
Extrolite profiles are fundamental taxonomic criteria in both filamentous and yeast fungi
T/F: Taq polymerase can proofread and correct misincorporated nucleotides during DNA amplification
FALSE
Taq polymerase lacks 3' → 5' exonuclease proofreading activity, so it cannot correct errors. High-fidelity polymerases like Pfu or Q5 have proofreading ability.
T/F: The length of the PCR product is equal to the distance between the two primers
FALSE
Amplicon length = target region L + F & R primers L
e.g., target region = 300 bp; F & R primers = 20 bp each
Amplicon length = 300 + 20 + 20 = 340 bp
Extrolite profiles are fundamental taxonomic criteria in filamentous and yeast fungi because _
the production of identified extrolites can be compared with known or published standards

T/F: In PCR, primers always bind to the template strand, never to the newly synthesized strand
FALSE
In later cycles, primers can bind to both template DNA and newly synthesized DNA strands, amplifying the product exponentially
T/F: A single PCR cycle produces double the number of DNA molecules as the previous cycle, making amplification purely linear
TRUE
A single PCR cycle produces double the number of DNA molecules as the previous cycle, making amplification exponential
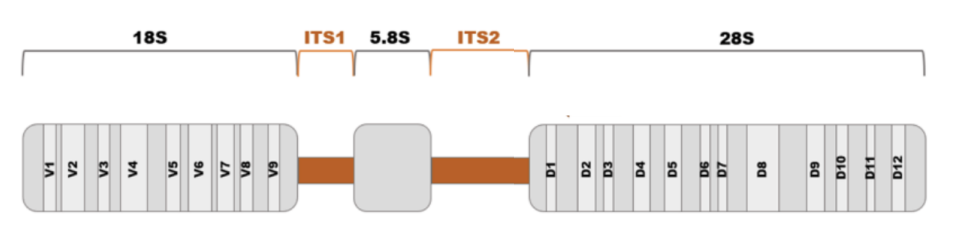
_ are common target regions for fungal DNA barcoding
28S LSU D1-D2
26S SSU
ITS region
Enumerate characteristics of a good molecular marker or target region
The marker should be universal, meaning it should be present in all fungi of interest to ensure applicability across different taxa
There should be sequence conservation for priming sites, i.e., the flanking regions of the target sequence should be highly conserved so that universal primers can consistently amplify that region across different species
At the same time, the internal sequence of the marker should have sufficient variation to distinguish between different species but still be conserved enough within a species
In a fungal ribosomal operon, there are 2 hypervariable internal transcribed spacer (ITS) regions, specifically _, and 3 conserved regions, specifically 18S, 5.8S, 28S, that contain variable domains, i.e., 9 for 18S and 12 for 28S
ITS1, ITS2

T/F: While 28S LSU D1-D2, 26S SSU, and ITS are the most common targets and thus work for most unknown samples, it is always better to use 2-3 target regions when working on invariable PCR results
TRUE
What are the common databases used for the identification of fungal sequences?
NIH NLM (National Library of Medicine) database
European Nucleotide Archive (ENA)
DNA Data Bank of Japan (DBBJ)
Unite community
Ribosomal Database Project (RDP)
*Your choice of database in identifying your PCR result can influence the identity you’ll see; hence, it’s always better to cross-check across different databases.
_ generates ions from a sample that are detected based on their time of flight
Matrix-Assisted Laser Desorption Ionization - Time-of-Flight Mass Spectrometry (MALDI-TOF MS)
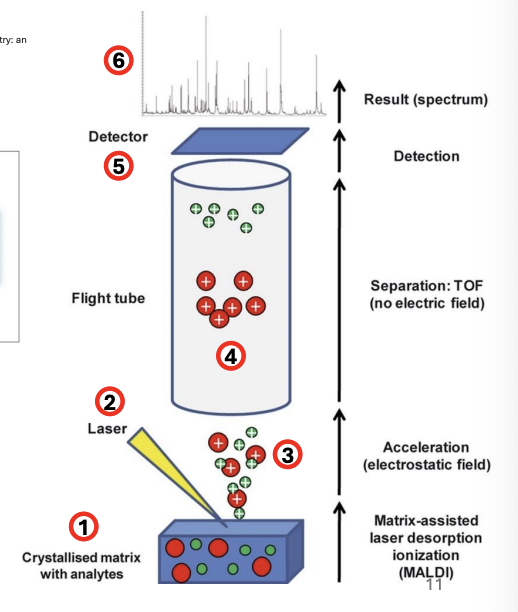
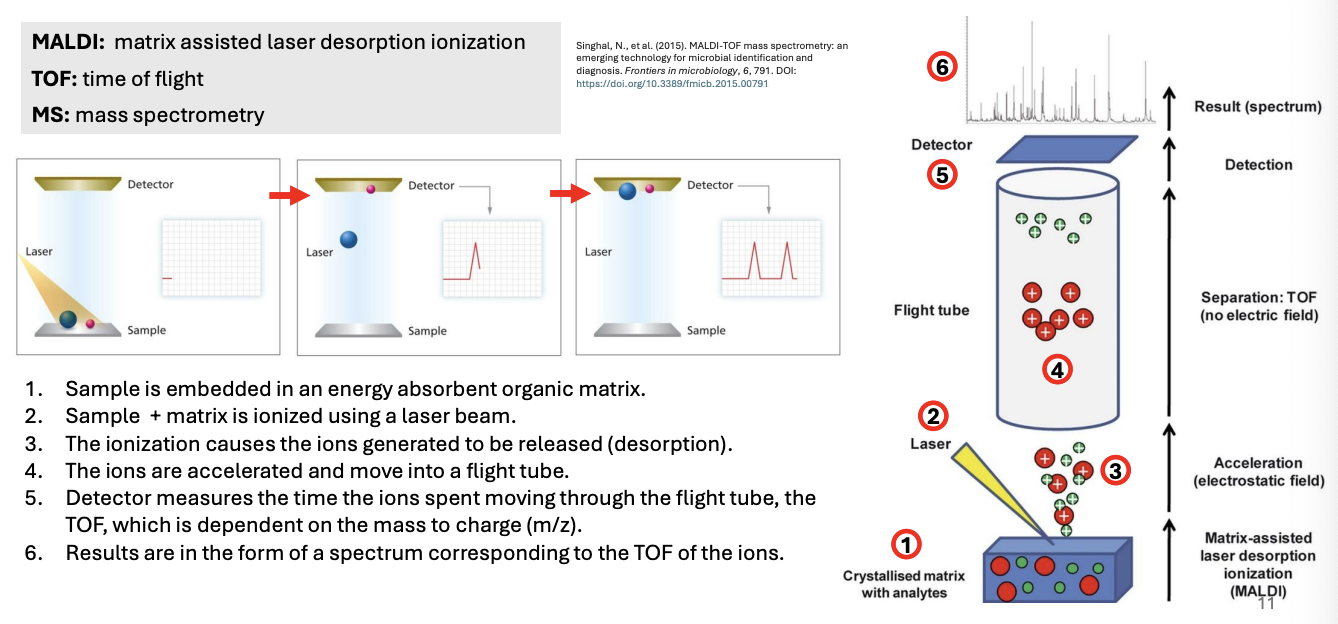
Explain MALDI-TOF as method for molecular identification of fungi
Sample is embedded in an energy-absorbent organic matrix
Sample + matrix is ionized using a laser beam
The ionization causes the ions generated to be released or desorbed (desorption)
The ions are accelerated and move into a flight tube
Detector then measures the time ions spent moving through the flight tube, i.e., time of flight, which is dependent on mass to charge ratio (m/z)
Results are in the form of a mass spectral chart that corresponds to the TOF of the ions
T/F: In MALDI-TOF, the matrix serves only to embed the sample and does not play a role in ionization
FALSE
The matrix is energy-absorbent and thus facilitates ionization by transferring energy from laser to sample, enabling desorption and ionization of analytes
T/F: The time of flight (TOF) of an ion is directly proportional to its mass-to-charge (m/z) ratio
FALSE
The time of flight (TOF) of an ion is directly proportional to the square root of its mass-to-charge (𝑚/𝑧) ratio, not to 𝑚/𝑧 itself
T/F: MALDI-TOF is a form of soft ionization, meaning it produces minimal fragmentation of biomolecules, making it ideal for microbial identification
TRUE
MALDI-TOF generates intact ions with minimal fragmentation, allowing the identification of characteristic protein mass patterns in microorganisms.
T/F: The laser in MALDI-TOF directly ionizes the sample molecules without any intermediary
FALSE
The laser primarily excites the organic matrix, which then transfers energy to the sample molecules, leading to their ionization and desorption.
T/F: Chytridiomycota do not possess chitinous cell walls at any trophic stage in their life cycle but have chitin in their dormant spores
FALSE
Cryptomycota (Rozellida) do not possess chitinous cell walls at any trophic stage in their life cycle but have chitin in their dormant spores
In a fungal ribosomal operon, there are 2 hypervariable internal transcribed spacer (ITS) regions, specifically ITS1 and ITS2, and 3 conserved regions, specifically _, that contain variable domains, i.e., _
18S, 5.8S, 28S
9 variable domains for 18S
12 variable domains for 28S
One of the symptoms of onychomycosis caused by Trichosporon mucoides is thick nails due to _ that it brings about
keratin hypersecretion
_ can differentiate between 2 species that cannot be separated through PCR
MALDI-TOF MS
Explain a case of MALDI-TOF MS resolving/delimiting species that cannot be separated through PCR
Trichosporon mucoides: a clinically important species that causes disseminated infections in humans, i.e., onychomycosis
Many T. dermatis strains have been mistakenly identified as T. mucoides through PCR
MALDI-TOF MS successfully identified the 2 species with 99.9% confidence value

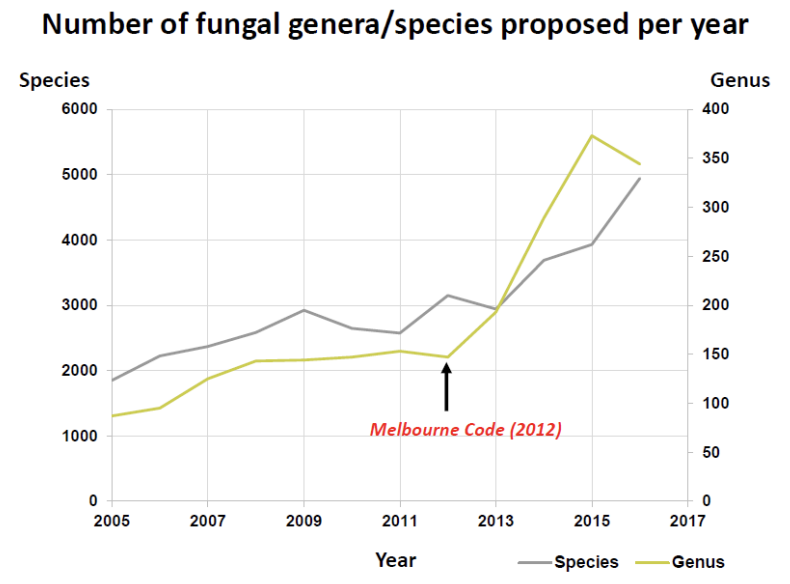
_ has caused a major change in fungal nomenclature, with the intent for each taxonomic group of fungi (taxon/taxa) to only have 1 correct name called “holomorph” that is accepted worldwide, provided that they have the same circumscription, position, and rank cpr
Melbourne Code of 2012 (Single Nomenclature; One fungus, One name = Holomorph)
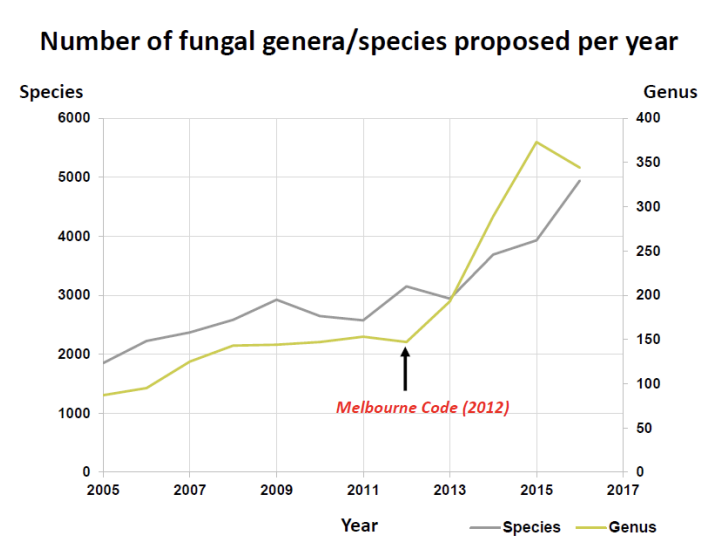
T/F: After the Melbourne Code of 2012, the proposed number of genera and species decreased
FALSE
It increased because differences in description and literature are now being consolidated and the use of different types of molecular barcodes and MALDI-TOF is separating what were once grouped together organisms
The most recent change in fungal taxonomy occurred last _, outlining the fungi and fungus-like taxa (Hyde et al., 530 authors)
Nov. 2024

What was the impact of the Single Nomenclature System (Holomorph, Melbourne Code of 2012) on fungal diversity estimates?
It led to an increase in the number of proposed species and genera because:
differences in literature and description are now being slowly consolidated and;
the use of molecular barcoding techniques and MALDI-TOF is separating what were once grouped together organisms

Explain Melbourne Code of 2012: Single Nomenclature (Holomorph = One name, One fungus); sample impacts
Goal was for each fungal taxonomic group to have only 1 correct name (holomorph) that is accepted worldwide, provided it has the same circumscription, position, and rank cpr
Back then, they had different names for the asexual and sexual forms of the same fungal species,
i.e., Anamorph = name for asexual form
i.e., Teleomorph = name for sexual form
"Dual names”
A = Candida; T = Pichia, Ogatea, Clavispora
A = Aspergillus; T = Eurotium, Emericella
A = Penicillium; T = Talaromyces, Eupenicillium
Holomorph (One name, One fungus)
Sample impacts
Penicillium (anamorph) and Talaromyces (teleomorph) are now 2 separate genera
Dissolution of Deuteromycota (fungi imperfecti) group
Dissolution of Zygomycota into several divisions (Mucoro-, Zoopagomycota)
_ have allowed species delimitation to become faster and more progressive
Molecular barcoding techniques, combined with morphological examination
T/F: In fungal identification, the ideal approach is to start with molecular data, followed by morphological analysis to confirm and refine identification
FALSE
In fungal identification, the ideal approach is to start with morphological data, followed by molecular analysis to confirm and refine identification
There are several types of organisms that constitute the fungi in a broad sense, including _
Kingdom Mycota (Fungi)
Probably derived from a choanoflagellate ancestor, 19 genera, focusing on 6 phyla ccb mab
Phylum Cryptomycota
Phylum Chytridiomycota
Phylum Blastocladiomycota
Phylum Mucoromycota
Phylum Ascomycota
Phylum Basidiomycota
Kingdom Straminapila
Derived from the protist group containing dog diatoms, oomycetes, golden brown algae loh
Labyrinthulomycota
Oomycota
Hyphochytridiomycota
Fungus-like organisms pdam
Plasmodiophoramycota (plasmodiophorids)
Dictyosteliomycota (dictyostelid slime molds)
Acrasiomycota (acrasid slime molds)
Myxomycota (plasmodial slime molds)

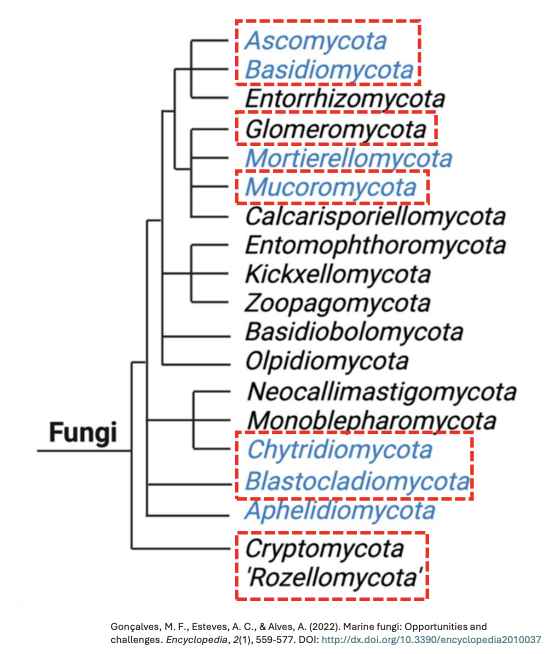
Majority of all fungi can be grouped into several _ which keep changing over time
major phyla or divisions
Which members of Kingdom Mycota have marine counterparts?
abm mcba
Asco-
Basidio-
Mortierello-
Mucoro-
Chytridio-
Blastocladio-
Aphelidio-
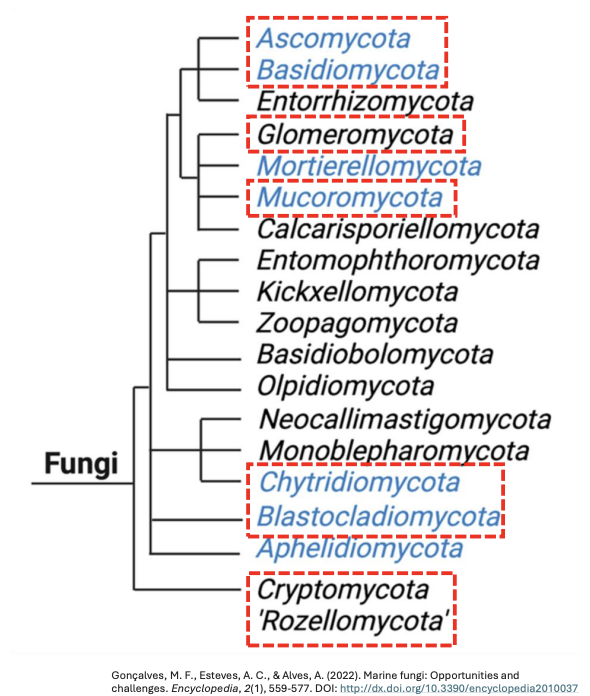
Explain Kingdom Mycota
ccb mab zg
Cryptomycota (Hidden fungi)
Differ from classic fungi due to their lack of chitinous cell walls
Most important class is Microsporidia, which is a group of spore-forming unicellular parasites sfup
Chytridiomycota
Most primitive of true fungi; most basal/unchanged
Mostly unicellular, some form hyphae
Gametes have flagella (zoosporic)
Blastocladiomycota
Originally under Chytridiomycota, zoosporic
Alternation of generations, massive diversity in form
Zygomycota (Conjugated fungi / bread molds)
Most are saprotrophic, form hyphae, produce resting spores = zygospores
Discontinued with members now part of Mucoromycota and Zoopagomycota (consists mostly of disease-causing fungi)
Mucoromycota (Mycorrhizae / bread molds)
Consists mainly of mrpm mycorrhizal fungi, root endophytes, plant decomposers, with some mycoparasites
Glomeromycota (Endomycorrhizae)
Small group, originally a phylum but now placed under Mucoromycota
Have hyphae but cannot survive outside roots
Ascomycota (Sac fungi)
Most known fungi belong to this group, have hyphae, produce sac called ascus containing ascospores (sexual spores)
Basidiomycota (Club fungi)
Have club-shaped cells called basidia for reproduction
Commonly form distinct fruiting bodies
_ do not possess chitinous cell walls at any trophic stage in their life cycle but have chitin in their dormant spores
Cryptomycota (Rozellida)

Describe the distribution of Cryptomycota
Fairly ubiquitous
Often not seen due to their microscopic nature
But detected due to their environmental DNA
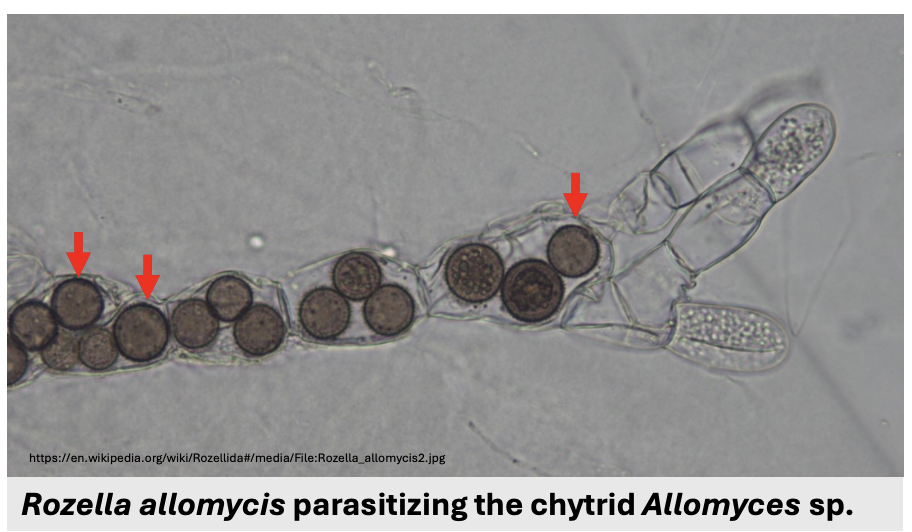
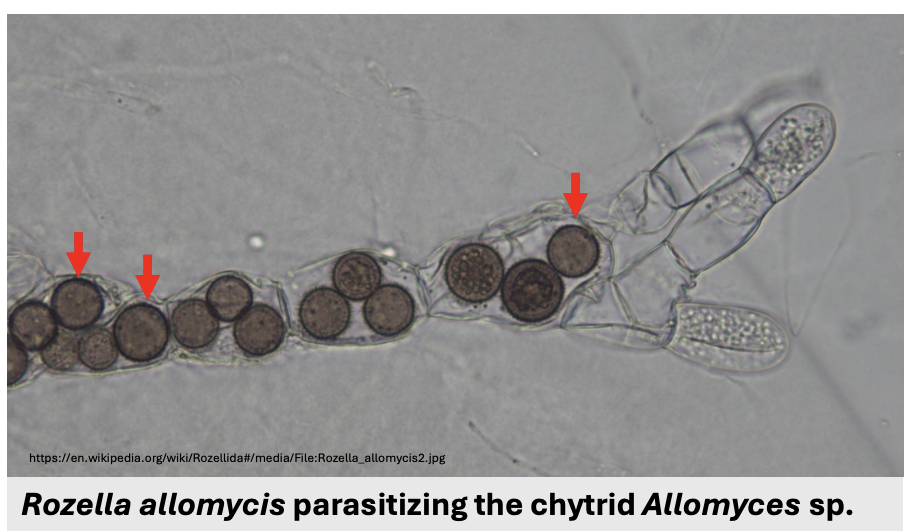
Since Cryptomycota do not have chitin in mature stages, they can be _ that feed by attaching to, engulfing, or living ael inside other cells
Describe figure shown
Phagotrophic parasites
Rozella allomycis parasitizing chytrid Allomyces sp.
Cryptomycota do not possess chitinous cell walls in their trophic life stages but have chitin in their _
dormant spores
Melbourne Code of 2012 caused a major change in fungal nomenclature with the intent for _
each taxonomic group of fungi to only have 1 correct name called “holomorph” that is accepted worldwide, provided that it has the same circumscription, position, and rank cpr
Microsporidia is a class under Cryptomycota that was originally under Protists but then reclassified under fungi because _
they’re closely related to the Chytrid fungi but obligate parasites of animals and protists

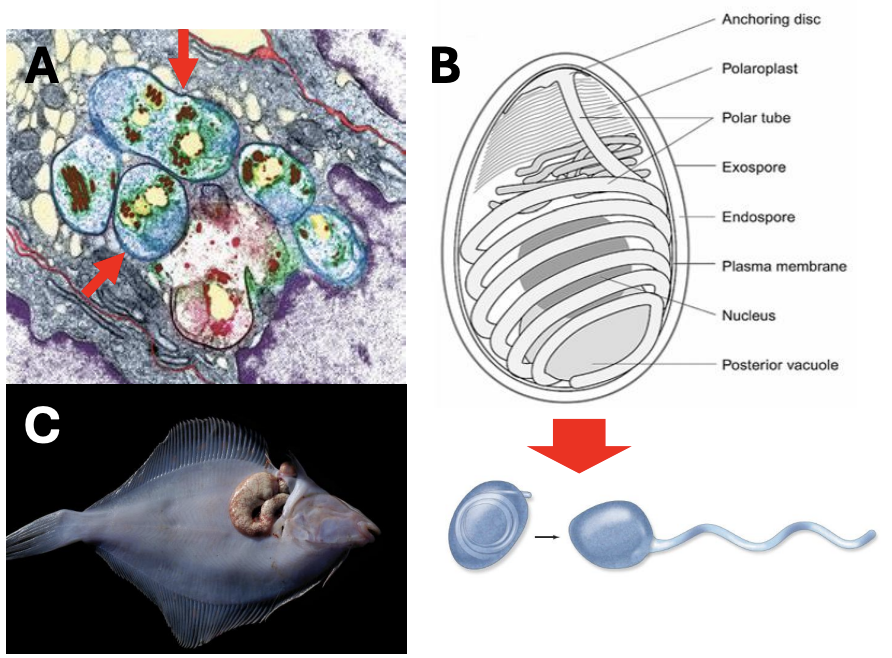
Describe the form of Microsporidia
They lack mitochondria and other organelles in many species
But they have a unique infection mechanism that uses a coiled polar tube inside its spores to penetrate and gain access to the host cell

Describe the ecology and reproduction of Microsporidia
Infection uses sporoplasm
Can be hyperparasites that infect parasites and form hypertrophic tissues called xenoma
T/F: Encephalitozoon proliferates freely in the cytosol, while Enterocytozoon proliferates within parasitophorous vacuoles
FALSE
Enterocytozoon proliferates freely in the cytosol (GI tract), while Encephalitozoon proliferates within parasitophorous vacuoles (lungs, GI)
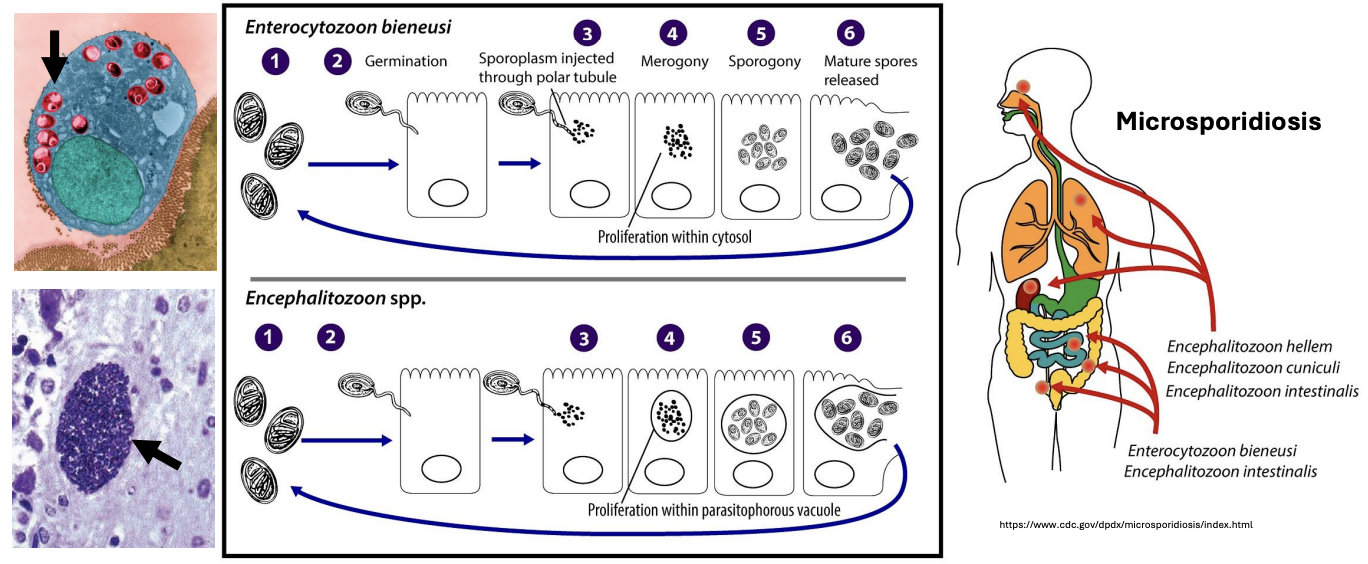
T/F: Once inside the host cell, Microsporidia lose their spore wall and do not re-form it until they exit the host
TRUE
The spore wall is shed after germination, and a new one is formed only during sporogony before release
_ has allowed better species delineation and description
Molecular barcoding
T/F: In the human body, Microsporidia mainly infect macrophages, allowing them to spread throughout different organ systems via the immune system
FALSE
They primarily infect epithelial cells, especially in the gut, though systemic infections can occur in immunocompromised individuals
Microsporidia produce _ which can persist for months and cause microsporidiosis
infective, resistant spores irs


Explain figure shown
Life cycle of 2 Microsporidia genera
*Microsporidia produce infective, resistant spores that can persist for months and cause microsporidiosis (affecting lungs, GI tract, even brain)
Enterocytozoon (GI tract) i gimsr
Infection begins with ingestion or inhalation of spores
Spores then germinate and
inject their sporoplasm into host cell polar tubules, eventually leading to
Merogony (asexual proliferation) in cytosol and
Sporogony (spore formation) in cytosol
Mature spores are then released to be able to infect other cells
Encephalitozoon (lungs, GI)
Infection via inhalation / ingestion
Germination
Injection of sporoplasm into host cell via polar tubules
Merogony within parasitophorous vacuole
Sporogony within parasitophorous vacuole
Release of mature spores to infect other cells
*Both respiratory and digestive systems are affected simultaneously whether infection started through inhalation or ingestion
_ are considered to be the earliest branch of true fungi
Chytrids


Chytrids are small, inconspicuous organisms that grow as _ on organic materials in moist soils or aquatic environments, _ no. of species, mostly aquatic
Single cells or primitively branched chains of cells
750 species
Distinguishing characteristic of Chytrids
They have motile, flagellated sexual zoospores mfsz


Many chytrids, e.g., Allomyces, form thallus anchored to substrates through _
rhizoids
Chytrids can be distinguished from one another through the _
structure of their zoospores, specifically their chitin composition
T/F: Microsporidia is a class under Cryptomycota consisting of spore-forming multicellular parasites
FALSE
Microsporidia is a class under Cryptomycota consisting of spore-forming unicellular parasites
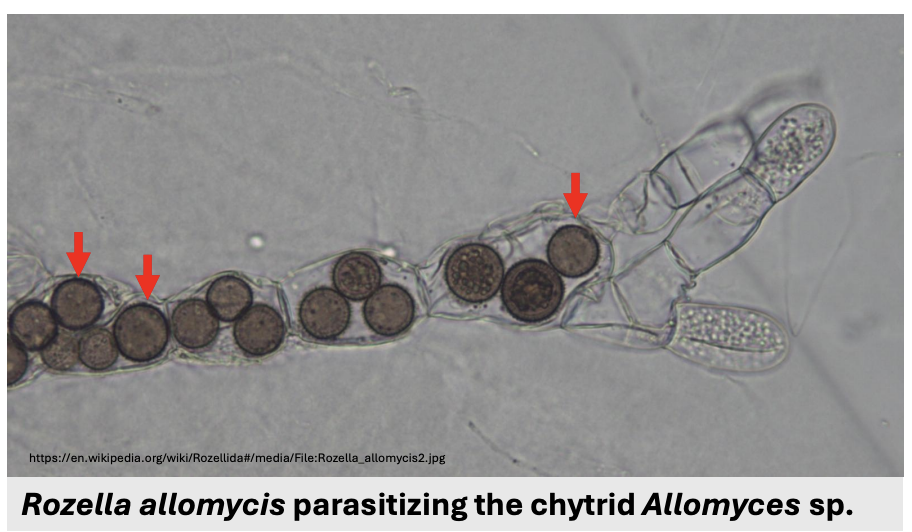
Since Cryptomycota do not chitin in mature stages, they can be phagotrophic parasites that feed by _
attaching to, engulfing, or living inside other cells ael
Microsporidia lack mitochondria and other organelles in many species but have unique infection mechanism that uses _ to penetrate and gain access to host cells
coiled polar tube inside its spores
Chytrids form _ for reproduction on various substrates
motile zoospores
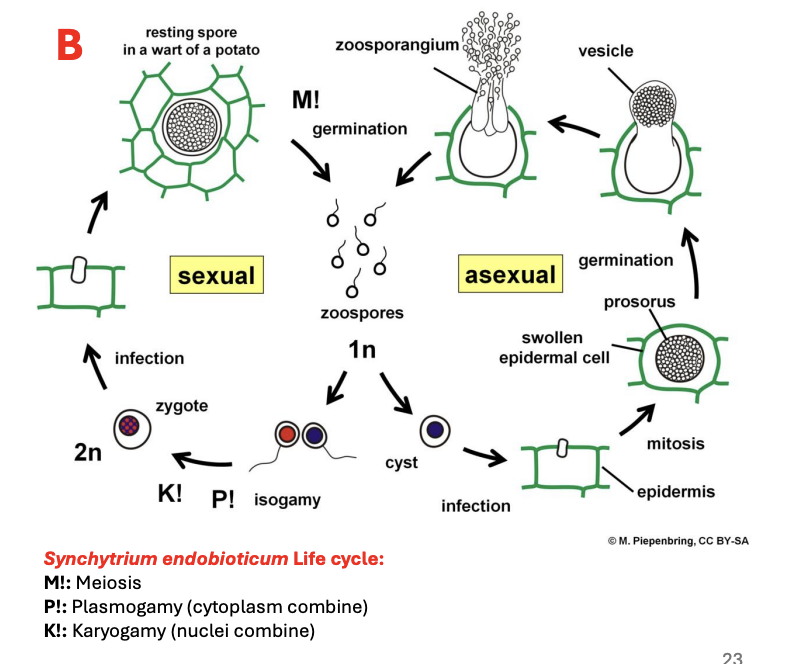
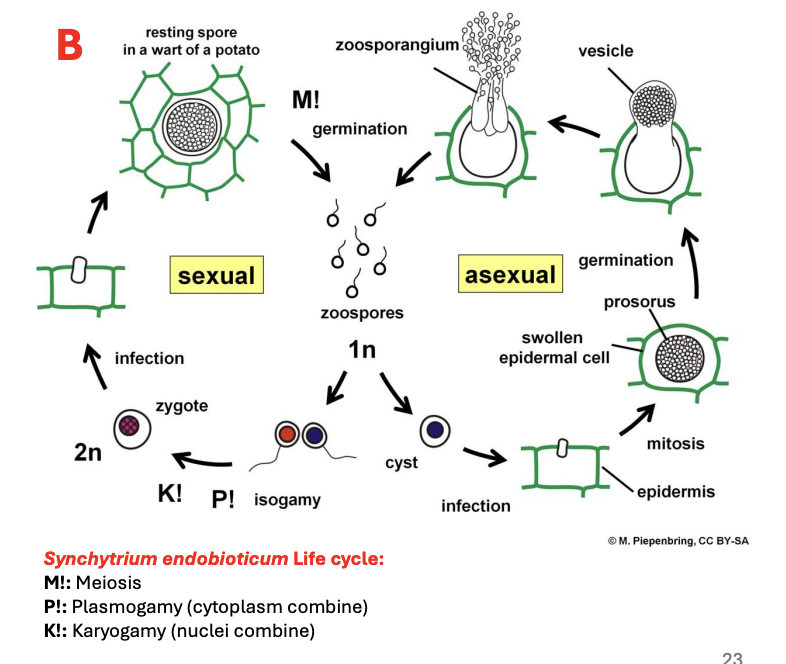
Describe ecology of Chytrids
Can be parasites of plants, animals, algae, and other fungal spores paaf
Batrachochytrium dendrobatidis causes chytridiomycosis of frogs
Describe reproduction of Chytrids
Reproduce mostly asexually
Sexual reproduction is observed and present
Zoospores = main method of dispersal and reproduction mmdr
Isogametes (i.e., gametes of the same size and shape) may be used

Microsporidia infect using _ and can be _ that infect parasites, forming hypertrophic tissues called xenoma
Sporoplasm
Hyperparasites