Lecture #13.2 | Oncogenes as Transcription Factors c-Myc and NFkB Ubiquitin and Proteasome Chronic Inflammation
1/15
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
16 Terms
c-myc
An Early Response Gene; does not need transcription to be activated
Activated by growth factors in serum.
Activity peaks at approximately 1-2 hours after serum addition.
c-Fos and c-Jun are also early response genes.
HOWEVER: slower, so not an immediate response gene
C-Myc is not detectable in quiescent fibroblasts (non-proliferating cells)
Characteristics of c-Myc
439 amino acids.
An early response gene.
A basic Helix-Loop-Helix (bHLH) protein.
Belongs to a family of TFs
Complex network of homo- and heterodimers
Binds DNA as a heterodimer with Max
Activates critical genes required for cell proliferation.
Activated by gene amplification, not mutation.
Synergistically induces tumors with Ras.
Plays a role in apoptosis and differentiation.
Myc/Max: basic Helix-Loop-Helix
B-basic region that interacts with DNA
helix makes interactions more flexible
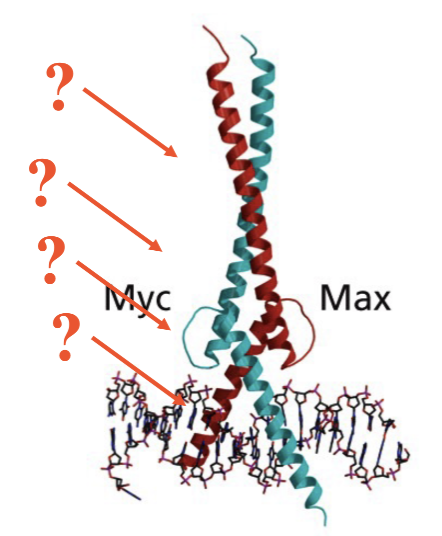
Functional domains of c-Myc and Max
Basic
HLH
Leu Zipper
Also NLS motif: nuclear localization sequence
helps localize to the nucleus as they cause the transcription themselves
Myc is longer due to the TAD: transactivation domain binds co-activators (only one of family that have)
Heterodimerization with Max
members of Myc/max family can all heterodimerize with Myc
Myc:Max heterodimers activate genes important for cell proliferation (e.g., cyclins, CDKs).
Max:Mxi, Max:Mad, and Max:Max all bind CACGTG elements but repress txn (only Myc has the TAD).
How can Myc:Max activate transcription and compete with Mad-Max
via recruitment of HATs and HDACs.
Myc can recruit HATs: Histone Acetyltransferases (cause acetylation of chromatin, loosening up DNA to make it more available for transcription)
MAD can recruit HDACs: Histone Deacetylases (cause deacetylation becoming more tightly bound)
Myc-Max and Mad-Max compete for regulation of target genes (cyclins, CDK) via binding E boxes.
Importance of Myc in forming tumors
Wild-type mouse embryonic stem (ES) cells form large tumors that become vascularized.
ES cells lacking c-Myc do not, showing c-Myc's essential role in tumor cell proliferation.
Lacking → much smaller tumors
How is c-Myc being activated into an oncogenes?
Gene amplification, chromosomal rearrangement, and amplification if the gene itself
myc is present on chromosome 8
Myc gets placed on chromosome for Immunoglobin H promoter, which is very active, resulting in Burkitt’s lymphoma.
Reciprocal translocation: the exchange of material between two chromosomes
Juxtaposition of the strong IgH promoter next to the c-Myc gene results in overexpression.
Insertion of a strong viral promoter next to c-Myc also leads to overexpression.
chromosomal rearrangement to make Myc oncogenic
myc is present on chromosome 8
Myc gets placed on chromosome for Immunoglobin H promoter, which is very active, resulting in Burkitt’s lymphoma.
Reciprocal translocation: the exchange of material between two chromosomes
Juxtaposition of the strong IgH promoter next to the c-Myc gene results in overexpression.
Insertion of a strong viral promoter next to c-Myc also leads to overexpression.
Activation of Myc through amplification of the gene itself
Amplifies the amount of DNA not just RNA and protein
through double minutes and homogeneous staining region
both lead to increased transcription of mic
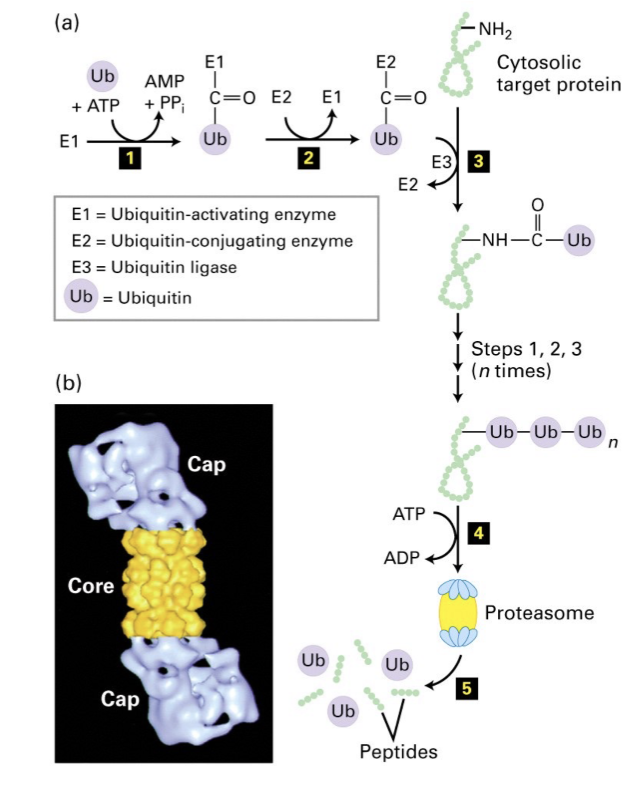
How are normal c-Myc protein levels maintained
Via ubiquitination (Ub = 76 aa polypeptide) of lysine residues that act as markers for the 26S proteasome pathway.
Three enzymes involved in ubiquitination:
E1: Ub activating enzyme
E2: Ub conjugating enzyme
E3: Ub ligase
the n-terminal domains
Macrophages
Phagocytic cells of the immune system.
They move, phagocytose bacteria, and engulf tumor cells.
They release inflammatory mediators:
Reactive oxygen species (ROS) and nitrogen oxide radicals
Act as initiators
Produced by enzymes COX2 and iNOS
Other enzymes:
MMPs: matrix metalloproteases, degrade ECM
Cytokines and chemokines:
TNFalpha
IL1
IL6
IL8
MCP1
Cancer creates increase of inflammatory mediators
Cytokines and chemokines trigger a signaling pathway that activates the txn factor NFkB
NFkB
A transcription factor (TF) and member of the Rel family.
v-Rel is a viral oncogene.
NFkB is located in the cytoplasm in an inactivated form
Bound to an inhibitor, IkB.
IkB is phosphorylated by IkB kinase (IKK) in response to cytokines.
Releases NFkB which can then translocate to the nucleus.
Activates the transcription of its target genes.
Plays a role in inflammation and innate immune response.
Also cell proliferation, apoptosis, and cell migration.
Is activated by cytokines in response to viral infections
Plays an important role in HIV.
Overexpressed in many tumors
e.g., mammary, gastric, and colon tumors.
Is the link between inflammation and cancer
NFkB Pathway
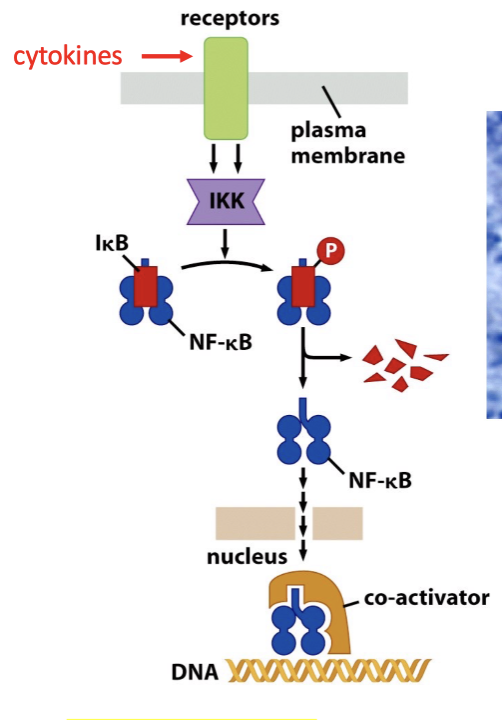
When IkB is phosphorylated, it disengages from NF-kB which is able to enter the cell and activate
can be used as a detection for early cancer development
What are the NF-kB target genes
Four classes of genes:
Autoregulation (IkB)
Inflammation (cytokines): Positive feedback loop
Anti-apoptosis
Proliferation (c-Myc & cyclin D1)
Role of NFkB in Gastric and Colon Cancer
H. pylori infection (ulcers), a risk factor for gastric cancer, activates the NFkB pathway.
NFkB turns on genes like COX2 and iNOS which generate ROS and RNS and promote inflammation and cause DNA damage.
It also turns on genes such as growth factors and cytokines that stimulate cell proliferation.
DNA damage and proliferation -- a lethal combination!!