4.3- Classification and Evolution (copy)
1/51
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
52 Terms
what is taxonomy
the practice of biological classification
why is classification required
allows us to arrange species into groups based on evolutionary origins and relationships
no overlap between groups→ each group called a taxon
makes it easier to understand and remember organisms
the hierarchal classification system
domain
kingdom
phylum
class
order
family
genus
species
the binomial naming system
universal way of identifying species
uses genus and species
italicised/underlined and
capital letter for Genus but not for species
three domains
bacteria
archaea
eukarya
archaea
often extremophiles
prokaryotes
different structure to bacteria
DNA transcription more similar to that of eukaryotes
bacteria
prokaryotes
divide by binary fission
eukarya
eukaryotes
divide by mitosis
can reproduce sexually or asexually
differences between bacteria and archaea
Feature | Bacteria | Archaea |
|---|---|---|
Membrane Lipids | unbranched hydrocarbon chains ester bonded to glycerol | branched hydrocarbon chains ether bonded to glycerol |
rRNA | base sequences more similar to rRNA of eukarya | |
Cell Wall Composition | peptidoglycan | don’t contain peptidoglycan |
the five kingdoms
highest rank of classification before three domains were introduced
prokaryota
Protoctista
fungi
plantae
animalia
prokaryota
bacteria and blue-green (photosynthetic) bacteria
unicellular
divide by binary fission
no nucleus
blue-green bacteria are autotrophic
most bacteria re heterotrophic
Protoctista
eukaryotic
unicellular or a group of similar cells
protozoa cells are similar to animal cells
algae cells are similar to plant cells
fungi
heterotrophic/ saprotrophic
can be unicellular e.g. yeast
some have long threads called hyphae which form mycelium network
some release spores that allow them to reproduce
plantae
multicellular eukaryotes
autotrophs
have complex body forms→ branching systems above and below ground
animalia
multicellular eukaryotes
heterotrophs
what is phylogeny
evolutionary history of organisms
show evolutionary relationships between taxa
sequence data used in phylogeny
DNA
mRNA
Amino Acids
DNA analysis and comparison
DNA extracted from nuclei of cells of an organism
base sequence obtained from extracted DNA
compared to that of other organisms to determine evolutionary relationships:
more similar base sequence= more closely related= more recent common ancestor
immunology in phylogeny
albumin protein found in many species and is commonly used for experiments
pure albumin samples extracted from blood of multiple species
each sample injected into a different rabbit
rabbit produces antibodies for specific type of albumin
antibodies extracted and mixed with different albumin samples
ppt from each mixed sample is weighed:
greater weight of ppt= greater complementarity between antibody and albumin
the work of Wallace and Darwin
Wallace collected specimens from south east Asia and South America
Darwin made several key observations:
not all offspring of an organism survive
populations of organisms fluctuate but not significantly
populations of the same species of organisms show variation between individuals
offspring inherit characteristics from their parents
wrote theory of evolution by natural selection
evidence for evolution by natural selection
fossils
molecular evidence
fossils
preserved remains of organisms or features left by organisms
fossils tell us that environments and organisms living in them have changed significantly over millions of years
fossils can be dated using carbon dating, so can be put into a sequence from oldest to youngest to see how organisms have changed through evolution
show similarities between extinct species, ancestral species and present day species
provides evidence for gradual change from simple life forms to complex life forms
molecular evidence for evolution by natural selection
DNA found in nucleus of cells can be sequenced and used to provide evidence of evolutionary relationships and how genetic code has changed as they have evolved
differences between nucleotide sequences in genes of species provides information:
more similar sequence= more closely related= species have separated more recently
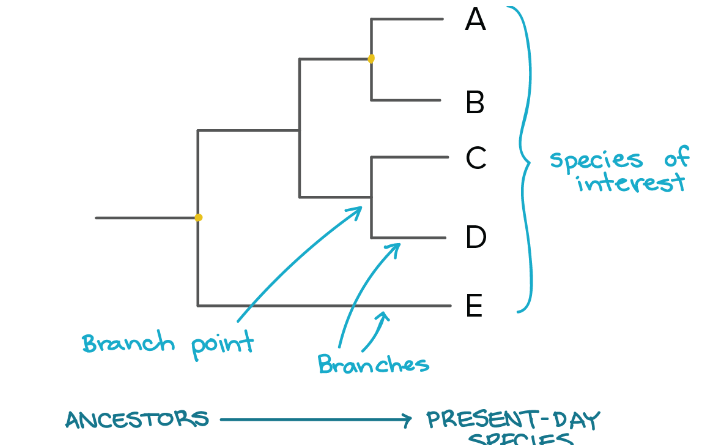
phylogenetic trees
show relationships between species
closer to end= more recent common ancestor
variation
differences that exist between organisms
can be:
genetic→ variation in genes
phenotypic→ variation in characteristics expressed
types of genetic variation
interspecific
intraspecific
interspecific variation
variation between species
useful in identifying different species
species that look v. similar can have forms of phenotypic variation that helps differentiate them
other species will have have slightly different niches that help distinguish between them
in cases where phenotypes are too similar, genotypes can be used to help identify them
intraspecific variation
variation within individuals of the same species
variation observed in phenotypes can be due to qualitative or quantitative differences
discontinuous variation
Caused by qualitative differences:
discrete and distinguishable categories e.g. blood groups
continuous variation
caused by quantitative differences:
range of values exist between two extremes e.g. height, mass
causes of variation
genetic factors
environmental factors
causes of discontinuous variation
occurs due to genetic factors- no environmental factors
different genes have diff effects on phenotypes
different alleles have large effect on phenotype
e.g. if earlobes are attached or free
causes of continuous variation
interaction between genetics and environment
genetic:
different alleles have small effect on phenotype
different genes can have same effect on phenotype
environmental factors:
length of sunlight hours
supply of nutrients
water availability
temp. range
oxygen levels
adaptation
features enabling organisms to survive in the conditions of their habitat
caused due to environmental factors giving rise to selection pressures→ natural selection
natural selection will select for favourable alleles that produce adaptations→ organisms with these adaptations will be more likely to survive and reproduce
types of adaptation
anatomical
physiological
behavioural
anatomical adaptations
structural or physical features
e.g. white fur of polar bear providing camouflage
physiological adaptations
biological processes within the organism
e.g. mosquitos produce chemicals so stop blood clotting
behavioural adaptations
the way an organism behaves
e.g. cold blooded reptiles bask in the sun
convergent evolution
species that do not share recent common ancestor can show high levels of similarity
occurs when two habitats are very similar→ organisms have similar adaptations
the process of natural selection
selection pressure exist in the environment due to environmental factors
random mutations produce new alleles of a gene
the new allele may benefit possessor, so there will be an increased chance of survival and increased reproductive success
advantageous allele is passed onto next generation
over several generations, new allele will increase in frequency in population
antibiotic resistance
within a bacterial population, there is variation caused by mutations
mutation causes some bacteria to become resistant to antibiotic
when population die, resistant bacteria do not die
resistant bacteria can reproduce with less competition from non-resistant bacteria
genes for antibiotic resistance are passed on w greater frequency to next generation
over time, whole pop. becomes resistant
how do bacteria inherit antibiotic resistance
vertical transmission
horizontal transmission
vertical transmission
bacteria reproduce asexually through binary fission
if one bacterium contains mutant gene, all of its descendants will carry antibiotic resistant gene
enables antibiotic resistance to spread within a population
horizontal transmission
plasmids contain antibiotic-resistant genes
plasmids frequently transferred between bacteria
occurs during conjugation:
tube forms between two bacteria to allow for exchange of DNA
bacteria containing mutant gene that gives antibiotic resistance can pass this gene on to other bacteria
enables antibiotic resistance to spread within or between populations
how to reduce cases of antibiotic resistance
only prescribe antibiotics when absolutely necessary→ not used in non-serious cases
ensure patients complete courses of antibiotics so all bacteria killed
only using antibiotics for bacterial infections
use highly specific antibiotics rather than wide spectrum antibiotics
rotate which antibiotics are used
hold some antibiotics back as a last resort
more investment into researching new antibiotics
selective agent
any environmental factor that influences the survival of a particular species and therefore drives natural selection in that species
consequences of antibiotic resistance
research into new antibiotics is time consuming and expensive
some strains of bacteria are resistant to multiple antibiotics→ infections are difficult to treat
can give rise to superbugs e.g. MRSA
commonly prescribed antibiotics are becoming less effective
bacteria can develop multiple resistance→ resistant to multiple antibiotics
consequences of pesticide resistance and how to reduce effects
consequences:
less food security as crops can still be ruined
mitigation:
use combination of pesticides
use sparingly or in rotation
using other forms of pest control e.g. biological control, genetic modification
Student’s t-test
used to determine if there is a significant difference between mean values of a particular variable across two populations
conditions:
data must be continuous and normally distributed
variances should be equal
samples must be independant
conducting a t-test
state null hypothesis→ assumes no significant difference between the means
calculate test statistic using t-test formula
calculate degrees of freedom:
n1+n2 -2
compare test statistic against critical value:
if t statistic>critical value, reject null hyp
if t statistic< critical value, accept null hyp
spearman’s rank correlation coefficient
used to measure strength and direction of association between two continuous variables that are not normally distributed
calculating spearman’s rank correlation
convert raw data values into ranks from smallest to largest value
if two values are the same, give them an average rank
calculate spearman’s rho
compare ρ to a critical value to determine significance of correlation:
close to +1= strong positive correlation
close to -1= strong negative correlation
close to 0= no correlation