SBI4U: U5 Gene Control & Molecular Techniques (Biotechnology)
1/128
Earn XP
Description and Tags
** Test Q's from 2022, *H- questions H mentioned
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
129 Terms
1. The "central dogma" of molecular biology
a. | explains the structural complexity of genes. |
b. | describes the flow of information. |
c. | is based upon the role of proteins in controlling life. |
d. | does not explain how genes function. |
e. | explains evolution in terms of molecular biology. |
b. | describes the flow of information. |
2. The genetic code
a. | is universal for all organisms. |
b. | is based on 64 codon, each with three nucleotides. |
c. | also comes equipped with punctuation marks. |
d. | is redundant. |
e. | all of these |
e. | all of these |
3. Molecules that interact with DNA to alter gene expression are
a. | operons. | d. | repressor amines. |
b. | promoters. | e. | carbohydrates. |
c. | regulatory proteins. |
c. | regulatory proteins. |
4. In negative control systems, which of the following would cause transcription to proceed?
a. | repressor | c. | cooperator | e. | DNA |
b. | operator | d. | inducer | ||
d. | inducer | ||
5. Repressor proteins
a. | prevent binding of RNA polymerase to DNA. |
b. | can be inactivated by an inducer such as lactose. |
c. | provide negative control. |
d. | prevent binding of RNA polymerase to DNA and can be inactivated by an inducer such as lactose. |
e. | prevent binding of RNA polymerase to DNA, can be inactivated by an inducer such as lactose, and provide negative control. |
e. | prevent binding of RNA polymerase to DNA, can be inactivated by an inducer such as lactose, and provide negative control. |
6. The lactose operon includes
a. | an operator. |
b. | three structural genes that manufacture lactose-metabolizing enzymes. |
c. | a promoter. |
d. | a repressor. |
e. | all but a repressor. |
e. | all but a repressor. |
7. The obvious advantage of the lactose operon system is that
a. | lactose is not needed as energy for bacteria. |
b. | lactose-metabolizing enzymes need not be made when lactose is not present. |
c. | the bacteria will make lactose only in the presence of the proper enzymes. |
d. | milk is not needed for adult humans' diet. |
e. | glucose can substitute for lactose in the diet of intolerant persons. |
b. | lactose-metabolizing enzymes need not be made when lactose is not present. |
8. The lactose operon
a. | requires the presence of milk in the environment of the bacteria. |
b. | is turned on before a baby is born. |
c. | is a control mechanism that enables vertebrates to digest milk. |
d. | causes the production of gas in the digestive tract of milk-drinking animals. |
e. | is an excellent model for explaining control mechanisms for eukaryotic forms. |
a. | requires the presence of milk in the environment of the bacteria. |
9. In prokaryotes, most of the control of gene expression is at the __________ level.
a. | transcriptional | d. | translational |
b. | transcript processing | e. | post-translational |
c. | transport |
a. | transcriptional |
10. Which of the following is part of a negative control mechanism?
a. | promoter |
b. | repressor |
c. | structural genes that produce lactase |
d. | operator |
e. | all of these |
b. | repressor |
11. The positive control of the lactose operon in bacteria is
a. | activated by a repressor protein. |
b. | independent of glucose concentrations. |
c. | activated by a protein known as CAP. |
d. | regulated by RNA polymerase. |
e. | all of these |
c. | activated by a protein known as CAP. |
12. In the prokaryote operon model, it is usually the __________ of a metabolic pathway that inactivates the repressor protein.
a. | enzyme | c. | substrate |
b. | product | d. | promoter |
c. | substrate |
13. When a gene transcription occurs, which of the following is produced?
a. | more DNA |
b. | protein or polypeptide sequences |
c. | messenger RNA |
d. | enzymes |
e. | genetic defects |
c. | messenger RNA |
14. Which of the following is NOT actually a part of an operon?
a. | promoter | d. | gene for acetylase |
b. | gene for permease | e. | regulator genes |
c. | operator |
e. | regulator genes |
15. A regulator gene produces which of the following?
a. | repressor protein | d. | operator |
b. | regulatory enzyme | e. | transcriber |
c. | promoter |
a. | repressor protein |
16. A repressor protein binds with
a. | messenger RNA. | d. | a product. |
b. | the operator. | e. | a substrate. |
c. | the regulator. |
b. | the operator. |
17. Which of the following is the region that is the binding site for RNA polymerase?
a. | heterogeneous DNA | d. | operator sequence |
b. | repressor gene | e. | all of these |
c. | promoter sequence |
c. | promoter sequence |
18. What is the name of the process in which a molecule combines with a repressor protein and changes the shape of the protein so that it no longer binds to the operator sequence?
a. | induction | c. | repression | e. | transcription |
b. | corepression | d. | translation | ||
a. | induction |
19. Four of the five answers listed below are features of the lactose operon. Select the exception.
a. | regulator | c. | operator | e. | structural gene |
b. | terminator | d. | promoter | ||
b. | terminator |
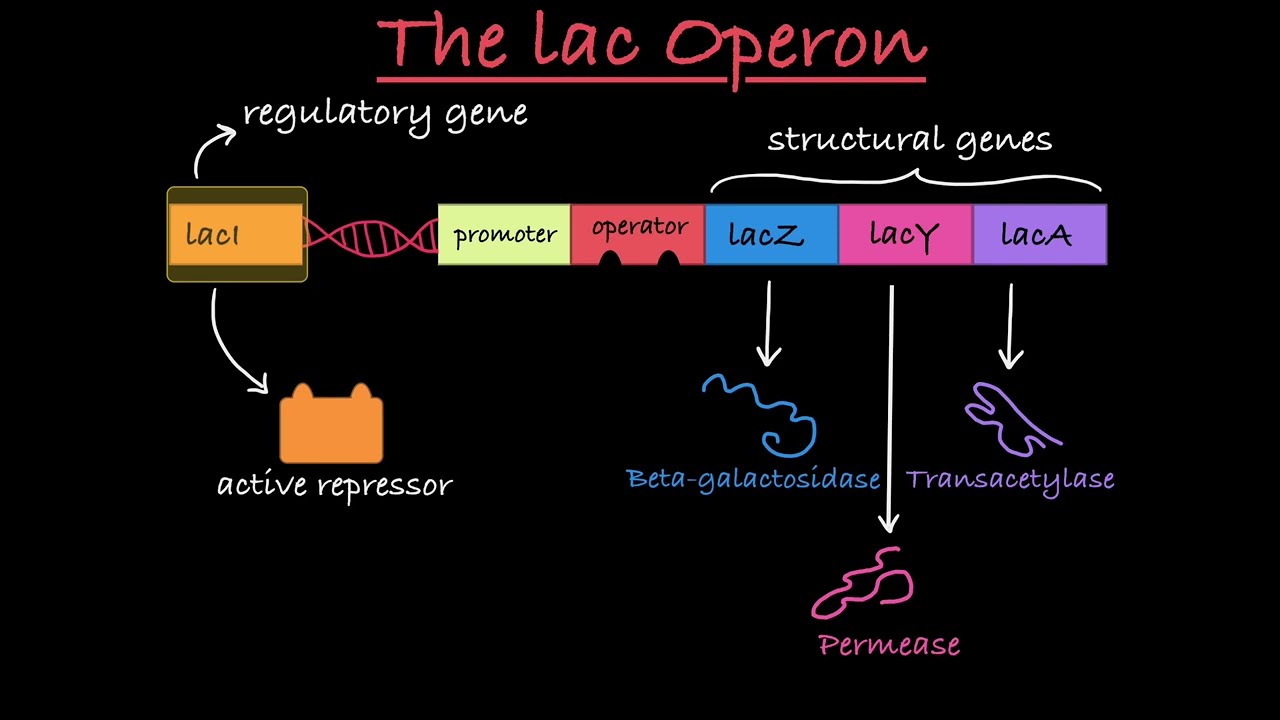
20. The nature of development (growth of an organism) is based upon
a. | which genes are active in a given cell. |
b. | when the genes function. |
c. | what gene products appear. |
d. | the amount of cellular products generated. |
e. | all of these |
e. | all of these |
21. Which of the following statements is FALSE?
a. | The patterns of gene expression vary from one cell to another in complex multicellular organisms. |
b. | Less is known about gene controls in unicellular prokaryotes than in multicellular eukaryotes. |
c. | Eukaryotes rely on conversion between active phosphorylated regulators and inactive forms more than prokaryotes do. |
d. | The timing of the induction (turning on) of genes varies throughout the life span of an organism depending upon the gene in question. |
e. | Hormones are one of the chief signaling molecules that allow for variable gene expression. |
b. | Less is known about gene controls in unicellular prokaryotes than in multicellular eukaryotes. |
22. Which of the following statements is FALSE?
a. | All the cells of any one single individual human have the same genes. |
b. | Only red blood cells have hemoglobin genes. |
c. | Different genes are activated depending on the cell they are in. |
d. | Cells don't express all of their genes all of the time. |
b. | Only red blood cells have hemoglobin genes. |
23. Genes located in different regions of the body during embryonic development may
a. | be turned on and off. |
b. | never be turned on. |
c. | be turned on and left on. |
d. | be activated for only a short time in one cell and a long time in another cell. |
e. | all of these |
b. | never be turned on. |
24. Which of the following terms refers to the processes by which cells with identical genotypes become structurally and functionally distinct from one another?
a. | metamorphosis | d. | differentiation |
b. | metastasis | e. | induction |
c. | cleavage |
e. | induction |
25. The control over the rate that mRNA reaching the cytoplasm is acted upon by ribosomes is known as
a. | transcriptional controls. |
b. | transcript processing controls. |
c. | transport control. |
d. | translational controls. |
e. | post-translational controls. |
d. | translational controls. |
26. The conversion of proinsulin into insulin by the removal of a portion of the polypeptide chain is an example of
a. | transcriptional control. |
b. | transcript processing control. |
c. | transport control. |
d. | translational control. |
e. | post-translational control. |
d. | translational control. |
27. Genes may be controlled by
a. | the way that they are packaged on a chromosome. |
b. | the X chromosomes in which they are located. |
c. | the dispersion of nucleosomes. |
d. | the production of more than one type of mRNA. |
e. | all of these |
e. | all of these |
28. X chromosome inactivation results in
a. | a total shut down of both female X chromosomes. |
b. | only the shut down of the X chromosome derived from the father. |
c. | only the shut down of the X chromosome derived from the mother. |
d. | the shut down of either the X from the father or the X from the mother. |
d. | the shut down of either the X from the father or the X from the mother. |
29. Eukaryotic nucleosomes exert control at what level?
a. | transcriptional | d. | translational |
b. | transcript processing | e. | post-transcriptional |
c. | transport |
d. | translational |
30. A mammalian female
a. | usually has one Barr body. |
b. | is a mosaic for the X-linked traits she inherits. |
c. | uses the paternal X chromosome for a Barr body. |
d. | uses the maternal X chromosome for a Barr body. |
e. | always has one Barr body and is a mosaic for the X-linked traits she inherits. |
a. | usually has one Barr body. |
31. There are a number of different small RNA molecules such as microRNA (miRNA) and small interfering RNA (siRNA). Both of these types of RNA are involved in control of gene expression by
a. | binding histones tightly to the DNA strand. |
b. | preventing RNApol from binding to the DNA. |
c. | acting as ‘road blocks’ that prevent translation. |
d. | preventing proper folding of the final protein. |
e. | removing the initial met a.a. from the protein. |
c. | acting as ‘road blocks’ that prevent translation. |
32. Which of the following is NOT a potential control mechanism for regulation of gene expression in eukaryotic organisms.
a. | the degradation of DNA. |
b. | the transport of DNA from the nucleus. |
c. | the lactose operon. |
d. | RNA interference. |
e. | Gene duplication. |
c. | the lactose operon. |
33. Four of the five answers listed below are types of gene control in eukaryotes. Select the exception.
a. | replicational control |
b. | transcriptional control |
c. | translational control |
d. | post-translational control |
e. | transcript processing control |
a. | replicational control |
34. Four of the five answers listed below are features exhibited by DNA and chromosomes. Select the exception.
a. | looped domain | d. | nucleosome |
b. | puff | e. | nucleolus |
c. | lampbrush |
e. | nucleolus |
35. Recombinant DNA
a. | has occurred in sexually reproducing forms. |
b. | can be produced with new biological techniques. |
c. | occurs with viral infections of various forms of life. |
d. | has produced changes that resulted in evolution. |
e. | all of these |
e. | all of these |
36. Genetic variations in populations are due to
a. | mutations. |
b. | sexual recombination. |
c. | crossing over. |
d. | genetic breeding programs. |
e. | all of these |
e. | all of these |
37. The goal of genetic engineering is to
a. | eliminate antibiotic resistance in bacteria. |
b. | isolate disease-causing plasmids. |
c. | transfer of viral genes. |
d. | modify cells to correct a defect or produce a desired product. |
d. | modify cells to correct a defect or produce a desired product. |
38. Recombinant DNA technology
a. | uses bacteria to make copies of the desired product. |
b. | splices DNAs together. |
c. | is possible only between closely related species. |
d. | uses bacteria to make copies of the desired product and splices DNAs together. |
e. | uses bacteria to make copies of the desired product, splices DNAs together, and is possible only between closely related species. |
d. | uses bacteria to make copies of the desired product and splices DNAs together. |
39. Small circular molecules of "extra" DNA in bacteria are called
a. | plasmids. | c. | pili. | e. | transferins. |
b. | desmids. | d. | F particles. | ||
a. | plasmids. |
40. Plasmids
a. | are self-reproducing circular molecules of DNA. |
b. | are sites for inserting genes for amplification. |
c. | may be transferred between different species of bacteria. |
d. | may confer the ability to donate genetic material when bacteria conjugate. |
e. | all of these |
e. | all of these |
41. After being transferred a plasmid must __________ to form a recombinant DNA molecule.
a. | be destroyed | d. | replicate |
b. | be integrated | e. | dissolve |
c. | undergo mutation |
b. | be integrated |
Integration in the context of molecular biology refers to the process by which a DNA molecule (like a plasmid or foreign DNA) becomes incorporated into the genome of a host organism.
42. Transformation
a. | is the direct transfer of DNA from one bacterium to another. |
b. | occurs when a bacterium acquires DNA from the surrounding medium. |
c. | is the result of crossing over. |
d. | occurs when a phage transfers bacterial DNA from one bacterium to another. |
e. | requires DNA polymerase. |
b. | occurs when a bacterium acquires DNA from the surrounding medium. |
43. In order for DNA molecules to undergo recombination they
a. | they must be from the same species. |
b. | their strands must separate as in replication. |
c. | must be cut and spliced at specific nucleotide sequences. |
d. | undergo lysis. |
e. | must be identical. |
c. | must be cut and spliced at specific nucleotide sequences. |
44. Enzymes used to cut genes in recombinant DNA research are
a. | ligases. | d. | DNA polymerases. |
b. | restriction enzymes. | e. | replicases. |
c. | transcriptases. |
b. | restriction enzymes. |
45. The "natural" use of restriction enzymes by bacteria is to
a. | integrate viral DNA. |
b. | destroy viral DNA. |
c. | repair "sticky ends." |
d. | copy the bacterial genes. |
e. | clone DNA. |
b. | destroy viral DNA. |
46. Which of the following is FALSE?
a. | Gene transfer and recombination is a common occurrence in nature. |
b. | The transfer of genetic material from one organism to another is dependent upon enzymes that cut and tie genes. |
c. | Rather than causing lysis of bacteria, some bacteriophages may be incorporated into the bacterial genome. |
d. | The insertion of gene fragments can be accomplished only in the laboratory under artificial conditions. |
e. | Once a gene has been incorporated into a bacterium, it may undergo amplification. |
d. | The insertion of gene fragments can be accomplished only in the laboratory under artificial conditions. |
47. The fragments of chromosomes split by restriction enzymes
a. | have fused ends. |
b. | have specific sequences of nucleotides. |
c. | have sticky ends. |
d. | form a circle. |
e. | both b) and c). |
f. | both b) and d) |
e | both b) and c). |
have specific sequences of nucleotides. |
have sticky ends. |
48. Restriction enzymes
a. | often produce staggered cuts in DNA that are useful in splicing genes. |
b. | are like most enzymes in being very specific in their action. |
c. | are natural defense mechanisms evolved in bacteria to guard against or counteract bacteriophages. |
d. | are used along with ligase and plasmids to produce a DNA library. |
e. | all of these |
e. | all of these |
49. Restriction enzymes
a. | work at recognition sites. |
b. | function only at "sticky ends." |
c. | produce uniform lengths of DNA. |
d. | function only in genetic laboratories. |
e. | none of these |
a. | work at recognition sites. |

50. Which enzyme was used to produce this molecule?
a. | ligase | c. | a restriction enzyme |
b. | transcriptase | d. | RNA polymerase |
c. | a restriction enzyme |
51. Which of the following enzymes joins the paired sticky ends of DNA fragments?
a. | reverse transcriptase | d. | DNA polymerase |
b. | restriction enzymes | e. | transferase |
c. | DNA ligase |
c. | DNA ligase |
52. Bacteria containing recombinant plasmids are often identified by which process?
a. | examining the cells with an electron microscope |
b. | using radioactive tracers to locate the plasmids |
c. | exposing the bacteria to an antibiotic that kills cells lacking the resistant plasmid |
d. | removing the DNA of all cells in a culture to see which cells have plasmids |
e. | producing antibodies specific for each bacterium containing a recombinant plasmid |
c. | exposing the bacteria to an antibiotic that kills cells lacking the resistant plasmid |
53. The method used to determine which host cells pick up a desired plasmid is the use of
a. | fluorescent dyes. |
b. | restriction enzymes. |
c. | antibiotics. |
d. | marker genes. |
e. | a known series of nonsense nucleotides (introns). |
c. | antibiotics. |
54. What is the most logical sequence of steps for splicing foreign DNA into a plasmid and inserting the plasmid into a bacterium?
I. Transform bacteria with recombinant DNA molecule.
II. Cut the plasmid DNA using restriction enzymes.
III. Extract plasmid DNA from bacterial cells.
IV. Hydrogen-bond the plasmid DNA to nonplasmid DNA fragments.
V. Use ligase to seal plasmid DNA to nonplasmid DNA.
a. | I, II, IV, III, V | d. | III, IV, V, I, II |
b. | II, III, V, IV, I | e. | IV, V, I, II, III |
c. | III, II, IV, V, I |
c. | III, II, IV, V, I |
III. Extract plasmid DNA from bacterial cells.
II. Cut the plasmid DNA using restriction enzymes.
IV. Hydrogen-bond the plasmid DNA to nonplasmid DNA fragments.
V. Use ligase to seal plasmid DNA to nonplasmid DNA.
I. Transform bacteria with recombinant DNA molecule.
Every Cat Hates Ugly Toads
55. Why are yeast cells frequently used as hosts for cloning?
a. | they easily form colonies |
b. | they can remove exons from mRNA. |
c. | they do not have plasmids. |
d. | they are eukaryotic cells |
e. | only yeast cells allow the gene to be cloned |
d. | they are eukaryotic cells |
56. The major advantage of using artificial chromosomes such as YACs and BACs for cloning genes is that
a. | plasmids are unable to replicate in cells. |
b. | only one copy of a plasmid can be present in any given cell, whereas many copies of a YAC or BAC can coexist in a single cell. |
c. | YACs and BACs can carry much larger DNA fragments than ordinary plasmids can. |
d. | YACs and BACs can be used to express proteins encoded by inserted genes, but plasmids cannot. |
e. | all of the above |
c. | YACs and BACs can carry much larger DNA fragments than ordinary plasmids can. |
57. Because it has no introns, researchers prefer to use __________ when working with human genes.
a. | cDNA | d. | RFLPs |
b. | cloned DNA | e. | viral DNA |
c. | hybridized DNA |
a. | cDNA |
58. A principal problem with inserting an unmodified mammalian gene into a bacterial plasmid, and then getting that gene expressed in bacteria, is that
a. | prokaryotes use a different genetic code from that of eukaryotes. |
b. | bacteria translate polycistronic messages only. |
c. | bacteria cannot remove eukaryotic introns. |
d. | bacterial RNA polymerase cannot make RNA complementary to mammalian DNA. |
e. | bacterial DNA is not found in a membrane-bounded nucleus and is therefore incompatible with mammalian DNA. |
c. | bacteria cannot remove eukaryotic introns. |
59. A gene that contains introns can be made shorter (but remain functional) for genetic engineering purposes by using
a. | RNA polymerase to transcribe the gene. |
b. | a restriction enzyme to cut the gene into shorter pieces. |
c. | reverse transcriptase to reconstruct the gene from its mRNA. |
d. | DNA polymerase to reconstruct the gene from its polypeptide product. |
e. | DNA ligase to put together fragments of the DNA that codes for a particular polypeptide. |
c. | reverse transcriptase to reconstruct the gene from its mRNA. |
60. Breaking the central dogma; RNA can manufacture DNA via the action of
a. | DNA polymerase. | d. | ligase. |
b. | RNA polymerase. | e. | restriction endonuclease. |
c. | reverse transcriptase. |
c. | reverse transcriptase. |
61. Which of the following methods of DNA amplification does NOT require cloning?
a. | reverse transcription |
b. | polymerase chain reaction |
c. | cloned DNA |
d. | reverse transcription and polymerase chain reaction. |
e. | polymerase chain reaction and cloned DNA. |
b. | polymerase chain reaction |
62. Which of the following best describes the complete sequence of steps occurring during every cycle of PCR?
1. | The primers hybridize to the target DNA. |
2. | The mixture is heated to a high temperature to denature the double stranded target DNA. |
3. | Fresh DNA polymerase is added. |
4. | DNA polymerase extends the primers to make a copy of the target DNA. |
a. | 2, 1, 4 | c. | 3, 4, 1, 2 | e. | 2, 3, 4 |
b. | 1, 3, 2, 4 | d. | 3, 4, 2 | ||
a. | 2, 1, 4 |
2. | The mixture is heated to a high temperature to denature the double stranded target DNA. |
1. | The primers hybridize to the target DNA. |
4. | DNA polymerase extends the primers to make a copy of the target DNA. |
63. For polymerase chain reaction to occur,
a. | isolated DNA molecules must be primed. |
b. | all DNA fragments must be identical. |
c. | the DNA must be separated into single strands. |
d. | a sticky end must be available for the ligase enzyme to function. |
e. | isolated DNA molecules must be primed and the DNA must be separated into single strands. |
e. | isolated DNA molecules must be primed and the DNA must be separated into single strands. |
64. The method used to produce single strands of DNA
a. | is known as amplification. |
b. | is heating a solution of DNA. |
c. | uses restriction enzymes. |
d. | isolates DNA molecules from the nucleus. |
b. | is heating a solution of DNA. |
65. Gel electrophoresis separates DNA molecules on the basis of
a. | the nucleotide sequence of their sticky ends. |
b. | their nucleotide sequences. |
c. | the amount of adenine they contain relative to the amount of thymine they contain. |
d. | the amount of adenine they contain relative to the amount of guanine they contain. |
e. | their lengths. |
e. | their lengths. |
66. Separation of DNA fragments by gel electrophoresis
a. | requires priming. |
b. | is controlled by the size of the fragment. |
c. | is based upon the negative charges of phosphate groups. |
d. | is easily accomplished. |
e. | all are correct except "requires priming." |
e. | all are correct except "requires priming." |
67. The use of RFLPs for "genetic fingerprinting" is based upon
a. | the type of gel used in electrophoresis. |
b. | identical alleles at loci. |
c. | differences of locations where enzymes make their cuts. |
d. | differences between blood and semen DNA. |
e. | bonding of DNA to RNA. |
c. | differences of locations where enzymes make their cuts. |
68. Which of the following statements about restriction fragment length polymorphism is false?
a. | RFLPs can be used as a genetic fingerprint. |
b. | RFLPs are based upon variations in alleles at the same locus. |
c. | RFLPs reflect the fact that molecular differences in alleles alter the site where restriction enzymes function. |
d. | RFLPs can be used to distinguish between identical twins. |
e. | RFLPs have greatly increased the number of sites involved in mapping the human genome. |
d. | RFLPs can be used to distinguish between identical twins. |
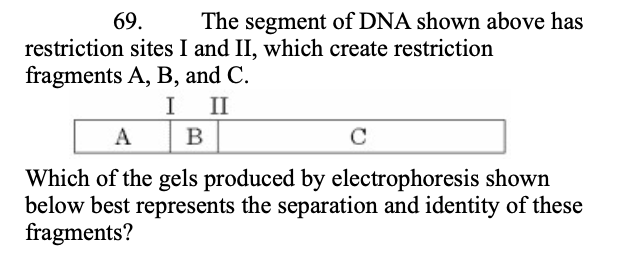
69. The segment of DNA shown above has restriction sites I and II, which create restriction fragments A, B, and C.
Which of the gels produced by electrophoresis shown below best represents the separation and identity of these fragments? (see image)
(see image for choices)
answer is B

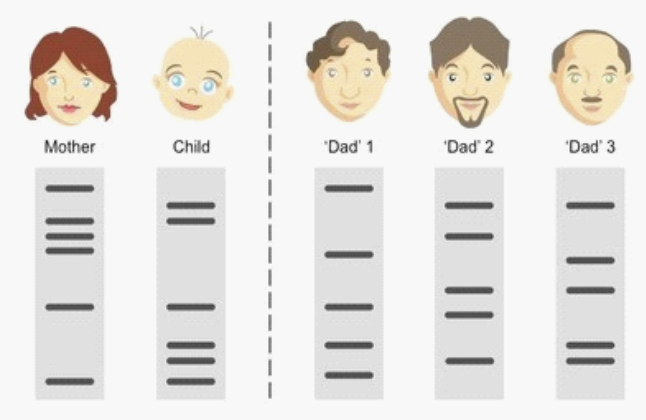
70. Who is the father?
a. | Dad 1 |
b. | Dad 2 |
c. | Dad 3. |
c. | Dad 3. |
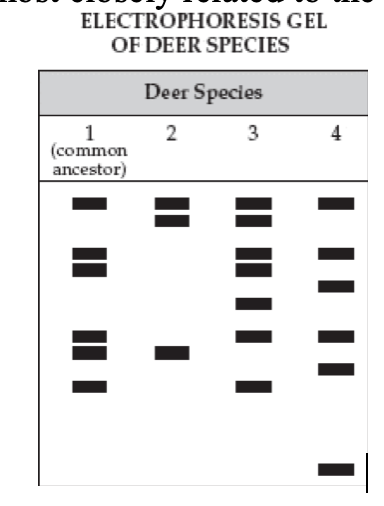
71. This gel shows the DNA of a common ancestor of deer species. Which of the three species is most closely related to the common ancestor?
a. | species 2 | c. | species 3 | e. | species 4 |
b. (im guessing its supposed to be c??) species 3
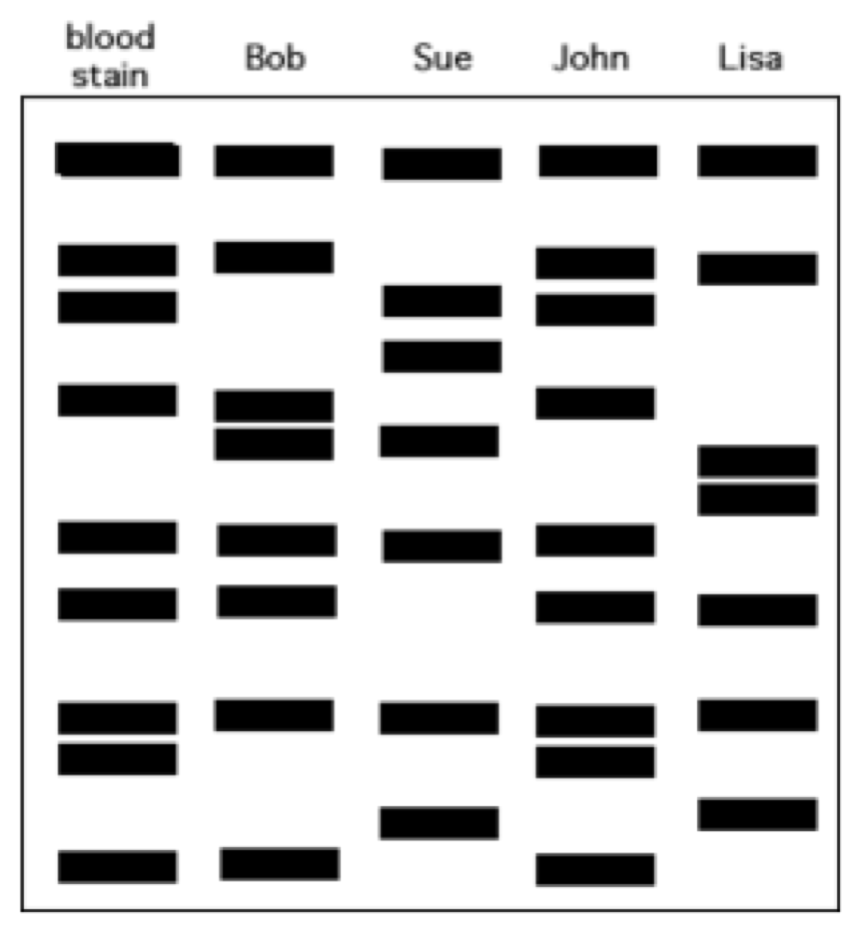
72. Bob was found dead in his home. Crime scene analysis showed signs of a struggle. Blood was collected from the crime scene, the victim and three suspects. Which suspect likely committed the crime?
a. | Sue | c. | John | e. | Lisa |
b. (again im assuming c?) John
73. Which was developed by a British researcher and causes DNA sequences to be transferred to a membrane and identified with a probe?
a. | Southern blotting | d. | Eastern blotting |
b. | Northern blotting | e. | RT-PCR |
c. | Western blotting |
a. | Southern blotting |
74. Probes for cloned genes use
a. | complementary nucleotide sequences labeled with radioactive isotopes. |
b. | certain media with specific antibodies. |
c. | specific enzymes. |
d. | certain bacteria sensitive to the genes. |
e. | all of these |
a. | complementary nucleotide sequences labeled with radioactive isotopes. |
75. The DNA fragments produced by automated DNA sequencing are identified using
a. | radioactive probes. | d. | electron microscopy. |
b. | laser beams. | e. | restriction enzymes. |
c. | ultracentrifugation. |
b. | laser beams. |
76. Which of the following tools of recombinant DNA technology is incorrectly paired with its use?
a. | restriction enzyme–production of RFLPs |
b. | DNA ligase–enzyme that cuts DNA, creating the sticky ends of restriction fragments |
c. | DNA polymerase–used in a polymerase chain reaction to amplify sections of DNA |
d. | reverse transcriptase–production of cDNA from mRNA |
e. | electrophoresis–separation of DNA fragments |
b. | DNA ligase–enzyme that cuts DNA, creating the sticky ends of restriction fragments |
77. A collection of DNA fragments produced by restriction enzymes and incorporated into plasmids is called
a. | copied DNA. | d. | a DNA library. |
b. | transcribed DNA. | e. | plasmid DNA. |
c. | DNA amplification. |
d. | a DNA library. |
78. Multiple copies of DNA can be produced by
a. | cloning a DNA library. |
b. | genetic amplification. |
c. | the use of reverse transcriptase. |
d. | the action of DNA polymerase. |
e. | all of these |
e. | all of these |
79. Which of the following can be modified genetically with the least consequences to its future survival?
a. | humans | ||||
b. | rats | ||||
c. | carrots | ||||
d. | bacteria | ||||
e. | plants | ||||
d. | bacteria | ||||
80. DNA microarrays have made a huge impact on genomic studies because they
a. | can be used to eliminate the function of any gene in the genome. |
b. | can be used to introduce entire genomes into bacterial cells. |
c. | allow the expression of many or even all of the genes in the genome to be compared at once. |
d. | allow physical maps of the genome to be assembled in a very short time. |
e. | dramatically enhance the efficiency of restriction enzymes. |
c. | allow the expression of many or even all of the genes in the genome to be compared at once. |
81. One of the first successful applications of genetic engineering was the commercial production of
a. | clotting factor. |
b. | insulin. |
c. | hemoglobin. |
d. | strawberries. |
e. | carrot seedlings. |
b. | insulin. |
82. Which of the following statements is FALSE?
a. | A plant has been genetically engineered with a bioluminescent gene from fireflies so that it glows in the dark. |
b. | Plants that have been generated from cultured cells derived from the same cell are identical clones. |
c. | Culturing of individual plant cells increases the mutation rates of their genes. |
d. | The insertion of genes into cultured plant cells uses the plasmid of a bacterial pathogen of plants. |
e. | Gene insertions are safer than pesticides because they are target-specific. |
b. | Plants that have been generated from cultured cells derived from the same cell are identical clones. |
83. What would be the value of inserting human genes suspected of causing Alzheimer disease into mice?
a. | to make mice feel more human |
b. | to make antibodies which can be injected into suffering patients |
c. | to develop mice that can serve as models for study of this condition |
d. | to use the mice for production of a vaccine |
c. | to develop mice that can serve as models for study of this condition |
84. The Human Genome Project
a. | identified the nucleotide sequence of all human genes. |
b. | developed a complete DNA library for a human gene. |
c. | developed genetic markers for all genetic diseases. |
d. | catalogued all the varieties of human alleles. |
e. | identified all humans that possess genetic defects. |
a. | identified the nucleotide sequence of all human genes. |
85. Which of the following statements is true?
a. | There is no danger involved in recombinant DNA research in humans. |
b. | There is no danger involved in recombinant DNA research in bacteria. |
c. | There is no danger in releasing recombinant organisms into the environment. |
d. | Stringent safety rules make the use of recombinant DNA research possible. |
e. | It is safe to conduct recombinant DNA research in plants. |
d. | Stringent safety rules make the use of recombinant DNA research possible. |
86. Gene therapy
a. | has not yet been used successfully with mammals. |
b. | is a surgical technique that separates chromosomes that have failed to segregate properly during meiosis II. |
c. | has been used successfully to treat victims of Huntington's disorder by removing the dominant damaging autosomal allele and replacing it with a harmless one. |
d. | replaces defective alleles with normal ones. |
e. | all of these |
d. | replaces defective alleles with normal ones. |
87. Four of the five enzymes below are used in genetic engineering. Select the exception.
a. | ligase |
b. | reverse transcriptase |
c. | restriction |
d. | replicase |
e. | DNA polymerase |
d. | replicase |
88. Four of the five statements below are true of cloned DNA. Select the exception.
a. | The plasmid used is the cloning vector. |
b. | Identical copies are produced. |
c. | Cloned DNA is produced by reverse transcriptase. |
d. | Multiple copies are produced. |
e. | Cloned DNA is manufactured in bacteria cells. |
c. | Cloned DNA is produced by reverse transcriptase. |
89. Dideoxyribonucleotide chain-termination is a method of
a. | cloning DNA. |
b. | sequencing DNA. |
c. | digesting DNA. |
d. | synthesizing DNA. |
e. | separating DNA fragments. |
b. | sequencing DNA. |
90. In 1997, Dolly the sheep was cloned. Which of the following processes was used?
a. | Use of mitochondrial DNA from adult female cells of another ewe. |
b. | Replication and dedifferentiation of adult stem cells from sheep bone marrow. |
c. | Separation of an early stage sheep blastula into separate cells, one of which was incubated in a surrogate ewe. |
d. | Insertion of a nucleus from an another adult sheep’s cell into an enucleated sheep egg, followed by incubation in a surrogate ewe. |
e. | Isolation of stem cells from a lamb embryo and production of a zygote equivalent. |
d. | Insertion of a nucleus from an another adult sheep’s cell into an enucleated sheep egg, followed by incubation in a surrogate ewe. |
91. As genetic technology makes testing for a wide variety of genotypes possible, which of the following is likely to be an increasingly troublesome issue?
a. | use of genotype information to provide positive identification of criminals |
b. | using technology to identify genes that cause criminal behaviors |
c. | the need to legislate for the protection of the privacy of genetic information |
d. | discrimination against certain racial groups because of major genetic differences |
c. | the need to legislate for the protection of the privacy of genetic information |
92. CRISPR is
a. | a family of DNA sequences derived from viruses in the genome of a bacterial host. |
b. | a gene editing tool. |
c. | an enzyme that can cut a specific matching target sequence. |
d. | a danger to the human race. |
b. | a gene editing tool. |
93. Which of the following best relates how CRISPR-Cas9 works?
a. | ‘measure and cut’ |
b. | ‘seek and destroy’ |
c. | ‘cut and paste’ |
d. | ‘trash and burn’ |
c. | ‘cut and paste’ |
94. What was the original function of the CRISPR system as it occurs in nature?
a. | Defending bacteria from viral infection. |
b. | Defending white blood cells from viruses. |
c. | Defending bacteriophages from bacteria. |
d. | Defending against bacterial infections. |
a. | Defending bacteria from viral infection. |
95. What possible risks are there in the modification of ‘designer babies’?
a. | New forms of illness may arise. |
b. | May remove certain genes ‘off-target’ that are important for the baby’s overall development and growth. |
c. | The baby gets super power mutations. |
d. | a) and b). |
d. | a) and b). |
a. | New forms of illness may arise. |
b. | May remove certain genes ‘off-target’ that are important for the baby’s overall development and growth. |
96. A PAM is
a. | a 15-20 bp sequence snipped from a viral genome that serves as a tracer DNA to locate viral DNA later on. |
b. | a popular girls name. |
c. | a 3-5 bp sequence that is adjacent to viral DNA in a CRISPR array. |
d. | the target sequence of Cas-9. |
d. | the target sequence of Cas-9. |
97. In the CRISPR/Cas-9 system, what is Cas9 and what does it do?
a. | an RNA molecule that binds to target DNA via complementary base pairing |
b. | a DNA sequence that binds the Cas9 protein |
c. | a viral protein that disrupts bacterial membranes |
d. | a protein enzyme that cuts both strands of DNA at sites specified by an RNA guide |
d. | a protein enzyme that cuts both strands of DNA at sites specified by an RNA guide |
98. An application of DNA technology to help environmental scientists would be
a. | use PCR to analyze DNA at a crime scene. |
b. | create a tobacco plant that glows in the dark. |
c. | clone the gene for human growth hormone to treat pituitary dwarfism. |
d. | make transgenic bacteria that can be used to clean up oil spills more quickly than do the natural bacteria. |
d. | make transgenic bacteria that can be used to clean up oil spills more quickly than do the natural bacteria. |
99. A GMO is an organism
a. | whose genes / DNA have been edited or altered. |
b. | that has received new stem cells in a bid to cure a disease. |
c. | whose entire genome has been sequenced. |
d. | whose DNA has been copied into a cDNA library. |
a. | whose genes / DNA have been edited or altered. |
100. Unlike for a somatic cell, an alteration in the germ line could potentially correct a genetic defect in
a. | and affected individual only. |
b. | an affected individual and his/her offspring. |
c. | an affected individual and all of his/her descendants.. |
d. | no one. The germ line cannot be altered. |
c. | an affected individual and all of his/her descendants.. |