BIOL 2010 - DNA damage and repair
1/20
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
21 Terms
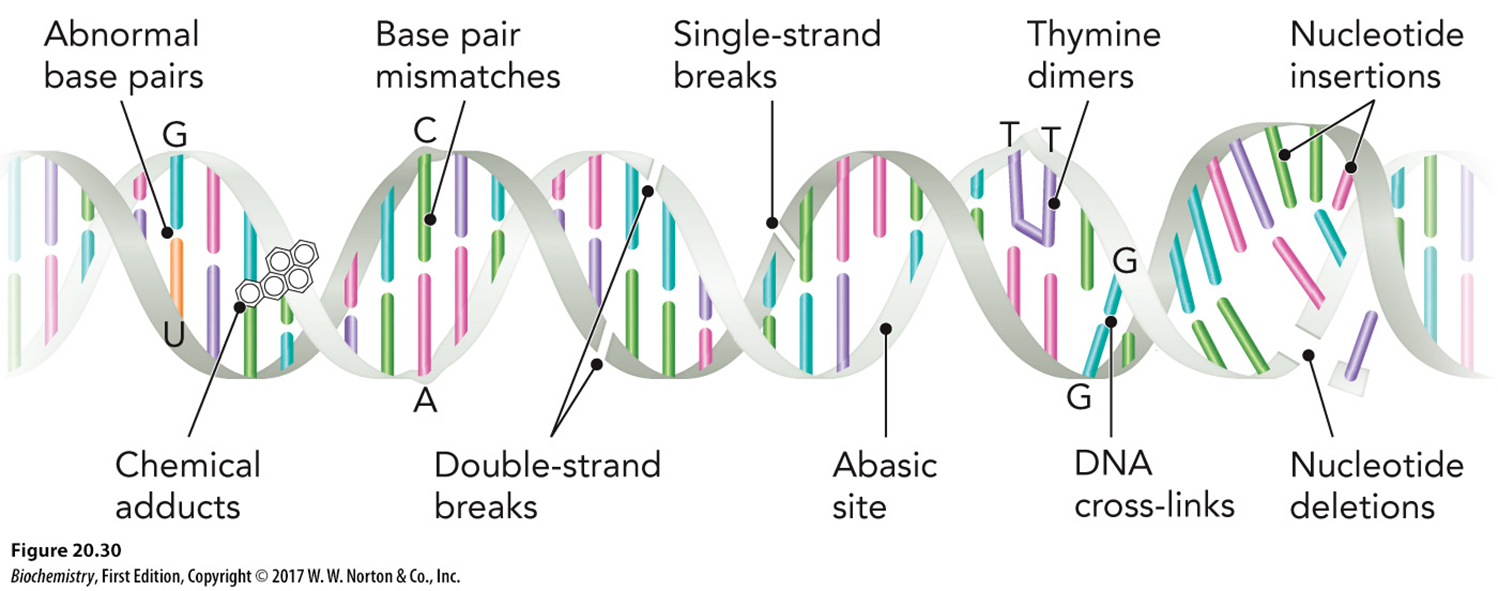
Types of DNA damage
abnormal base pairs
chemical adducts —> stop replication fork/DNA pol
incorrect bps
double stranded break —> fragments may be lost
single stranded break
abasic site —> nucleotide still there but no base
thymine dimers —> DNA pol cant get past
DNA cross links —> covalent bonds instead of H bonds
nucleotide insertions/deletions
spontaneous vs induced causes of DNA damage
SPONTANEOUS
replication errors
tautomerisation —> the addition of a proton at one molecular site and the removal of a proton at another
deamination
depurination
INDUCED
intercalating agents
base analogues
deaminating agents
oxidising agents
radiation, UV
Deamination/depurination
DEAMINATION
deamination of CAG can be repaired but deamination of T cant be repaired as its not considered wrong
deaminating agents will work much faster than spontaneous deamination
Guanine is much more prone to mutation than other bases as it has more sites of attack
DEPURINATION
depurination will not result in mutation until it becomes a template strand
4x more likely in ssDNA
AMES test
bacteria are modified so that they require histidine so only grow if histidine is present
can test if something is a mutagen by testing if the bacteria can grow on an agar plate without histidine
the mutagen will cause a mutation in the gene which allows the bacteria to produce histidine
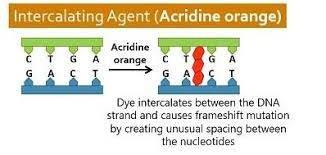
intercalating agents
fit between the tacked bases of DNA and change the width of the DNA strand
leads to distortion of the helix and local unwinding
can lead to slippage or improper base pairing which can cause frameshift mutations
E.g ethidium bromide
base analogues
molecules which resemble nucleotides and insert themselves into DNA but have altered pairing properties
E.g 5 bromouracil

oxidising agents
highly reactive oxygen species derived from by-products of cellular reactions
can cause strand breaking or cross links in the DNA
E.g hydroxyl radicals
radiation/UV
radiation can induce chemical changes in the DNA which alter its structure
radiation can interact with surrounding molecules to produce reactive species which can interact with the DNA
UV causes the formation of thymine dimers - its not a mutation but clogs up the machinery for replication
DNA repair mechanism
2 broad repair mechanisms :
strip out and resynthesise
direct repair to DNA
double stranded breaks
is the most serious form of DNA damage and has two repair methods:
HOMOLOGOUR RECOMBINATION
fragments of damaged chromosome can be aligned and cross over with its homologue
forms two holliday junctions (one for each type of the break)
DNA synthesis and ligation correct the break
NON HOMOLOGOUR END JOINING
Kn proteins bind the two broken ends of the duplex fragments and then to eachother
DNA ligase IV recruited to the join
Process is not precise - may have overhanging ss
ligates both strands at once but some loss of nt
risk of deletion mutation
risk is that joins any strand of DNA —> if theres multiple breaks chromosome fixed in wrong order
p53 tumor supressor
detects damaged DNA
arrests the cell cycle —> gives DNA the time to repair before S phase
activates DNA repair
if damage is too severe activates apoptosis or sinescence
most tumprs have mutated p53
4 types of sequence repair
direct repair
base excision repair
nucleotide excision repair
mismatch repair
DIRECT REPAIR
damage is modified for normal bases
can convert back to normal nucleotide
E.g: methylation, removal of thymine dimers
simple correction
DEMETHYLATION
methyltransferase enzyme
transfers methyl group onto itself
sacrificial —> permenant modification of enzyme
THYMINE DIMERS
corrected with light and enzymes
DNA photolyase absorbs blue light and breaks T-T internucleotide bonds using FADH —> 2xTs restored
instant repair
mammals cant do this, must use other repair mechanisms
4 types of sequence repair
direct repair
base excision repair
nucleotide excision repair
mismatch repair
BASE EXCISION REPAIR
removal and replacement of bases
>6 DNA glycosylases which recognise abnormal bases and cleave them from the deoxyribose creating an abasic site
can deal with:
deaminated A/C methyl C
oxidised
alkylated
open ring
double bond loss
glycosylases flip out bases for closer inspection
if the base is incorrect, cleaves the base but the enzyme remains attached to label it for the next stage
This is done UDGase - the same enzyme which recognises the U in the lagging strand
this makes it difficult to distinguish U and T but the steric clash of the methyl group on the T is enough to distinguish it as correct
when Us are present in DNA
U suggests a mutation
base is removed by glycosylase —> baseless nt
separate protein is required for baseless nucleotides
AP (apyrimidinic) endonuclease will close the phosphodiester backbone - this will also work in deourination
in bacteria pol 1 nick translation restores T and DNA ligase seals the nick
in eukaryotes pol beta does this
4 types of sequence repair
direct repair
base excision repair
nucleotide excision repair
mismatch repair
NT EXCISION REPAIR
removal of a ss sequence of DNA
triggered by distorted DNA
can correct any kind of damage
mammals use this to remove thymine dimers (prokaryotes will use DNA photolyase)
UVr ABC exinuclease in E.coli removes sections of DNA
UVrB heterodimer scans DNA and binds distortions
once bound the 2x UVrA proteins lost and UVrC is recruited to the site
UVrB and UVrC slide away from each other (on the undamaged strand) and cut either side of the damaged section
specialised helicase UVrD then knocks off the fragment
4 types of sequence repair
direct repair
base excision repair
nucleotide excision repair
mismatch repair
MISMATCH REPAIR
detects incorrect combo of bases
mismatch distorts the helix which is detected by proteins
the incorrect base is then removed
the most newly synthesised DNA will be corrected as its more likely to be mutated
hemimethylated DNA provides info on which is the most newly synthesised strand
IN PROKARYOTES
MutH binds to unmethylated daughter strand at the origin
MutS binds to distorted region
Mut L binds to Mut S and brings it to Mut H
Mut H cleaves the daughter strand at the origin and removes all of the DNA up to the mistake
UVrD removes the daughter strand a little bit past the distortion
pol 3 will then fill in the missing nts and DNA ligase will fill the nick
IN EUKARYOTES
we have several homologues
MutL 1-5
Mut S 1- 6
no homologues of Mut H as out DNA doesn’t use hemi methylation
multipronged approach - specific G mutation
8 oxo G is a specific G mutation
Mut T recognises 8 oxo GTP and hydrolyses it so it can repair mutation before it even ends up in DNA
Mut M recognises * oxo G in DNA and removes it via BER —> leaves an abasic site
Mut Y recognises 8 oxo G opposite an A in DNA and removes the V via BER
Gs get damaged easily so specific repair mechanism for mutated Gs
error rate of polymerases
10^-5
with proofreading = 10^-7
with the repair mechanism = 10^-10
translesion synthesis
creates a problem when DNA replication is stopped due to incorrect or distorted nucleotides which might cause mutation
some polymerases add nts where processive, proofreading polymerases cant
after, the mutations can be corrected
pol IV and pol V are translesion synthesis pols which are not too fussy about matching nts —> bypass a blockage so replication is finished
xeroderma pigmentosum
cant repair UV damage
skin very sensitive to UV light —> 1000 fold increased risk of skin cancer
due to inherited defects in one of 8 distinct genes responsible for components of the NER complex
system defective everywhere but only skin exposed to UV light so defects only shown in skin
HNPCC (colon cancer)
mutation in homologues of mismatch repair (Mut S/Mut L)
accumulation of mutations in genome which lead to colon cancer