Handout 11 - General Aspects of Active Sites
1/15
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
16 Terms
What are the features of active sites?
3D cleft/crevice formed by groups that come from different parts of the aa sequence
Relatively small portion of the total volume of the enzyme
Unique microenvironment (I.e - water is often excluded unless it is a reactant, influences electrostatic intermediate; microenvironment optimal for job that needs to be done)
Substrates bind by multiple weak attractions that are reversible
Specificity of binding depends on the precisely defined arrangement of atoms in an active site; needs good complementarity between active sites and substrate (TS)
What are the two different views/general features of active sites?
Lock and Key
Induced fit
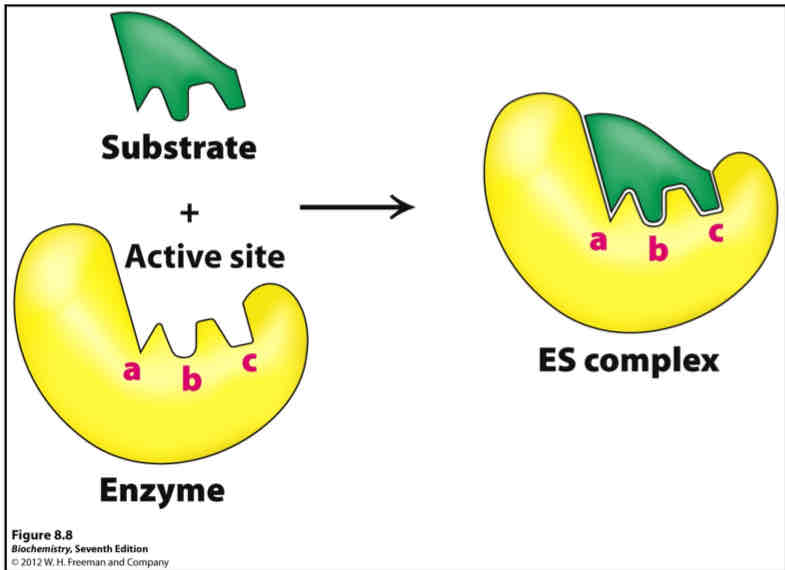
What is Lock and Key?
Pre-formed ligand-binding sites that are complementary in shape to their ligand (poor view of binding; simplistic model)
2 parts
Positioning or binding area (complementary to the substrate)
Catalytic area (R-groups + prosthetic groups interact with substrate to catalyze reaction)
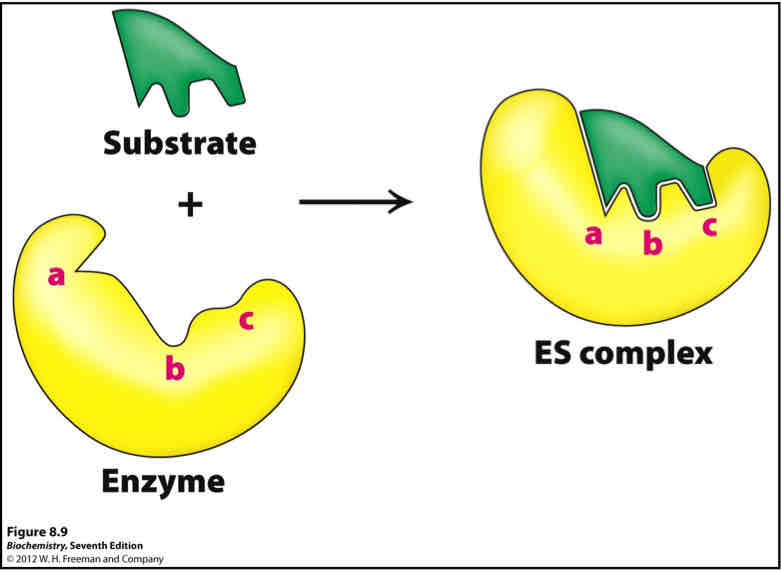
What is Induced fit?
Protein does not have optimal complementary at the binding site in the absence of ligand/substrate
Ligand induces a conformational change at the binding site that results in the complementary interaction
What are the 4 ways to probe an active site (gain info on chemistry of active site)?
Substrate analogs (look at km)
Competitive Inhibitors (look at ki)
Covalent Modification Agent (irreversible inhibition)
Site directed mutagenesis
What are substrate analogs?
Look at km
Measuring enzyme activity in the presence of different substrates
Looking to see if small change in substrate influences km
Km goes up = structural difference made substrate bind more weakly
Km goes down = structural difference made substrate bind better
What are competitive inhibitors (in regards to active site probing)?
Look at ki
Compete with substrate for binding at the active site; but the inhibitors are not chemically changed upon binding.
Ki = disassociation constant for EI complex
Smaller ki for given inhibitor = stronger binding/better complementarity
Bigger ki = weaker binding/worse complementarity
What are covalent modification agents (irreversible inhibition)?
Simple tools that react with specific R-groups
some specific
some react with small subset of available R-groups from aa’s
Simple: aa R-group specific
More Specific: Affinity Labels (Trojan Horses)
more specific to active site of a given enzyme
tricks enzymes to binding to active site
Swoops in for kill by covalently modifying the active site to kill activity
What is site directed mutagenesis?
Create mutant forms of the enzyme with aa swaps at key positions
evaluate performance of mutant relative to control wild type enzyme
Do each of these experiments for active site probing give the whole picture of the active site?
No, just little bits of information.
How is covalent modification used?
Covalent inactivation
treat enzyme with an agent specific for a particular aa
assess change in activity
decrease in activity = agent reacted with key aa at active site
Track down location of modification with peptide maps or mass spec
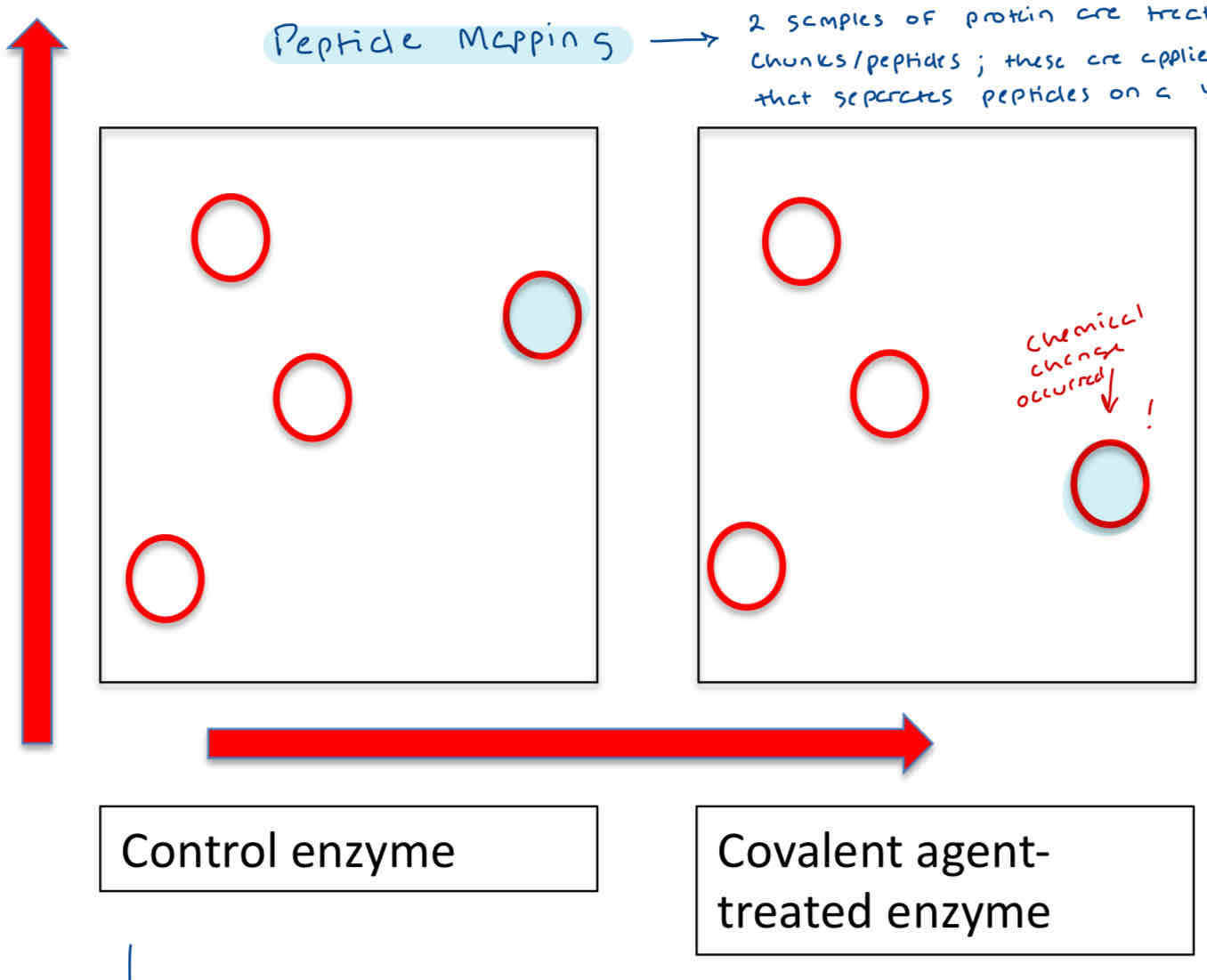
What is Peptide Mapping?
Old style method of separating the fragment on some sort of 2D chromatographic system (with different separation basis in each dimension)
polarity, charge, etc
Tells us which specific aa residue (i.e - the kind, position in sequence) was modified.
only small information about active site
Sequencing only the peptide fragments that showed a change in migration.
How is Affinity Labeling (aka Trojan Horse Inhibitors) used?
Substrate analog (part of structure that looks like substrate) bearing a covalent modification (electrophile) agent to specifically guide the modification agent to the active site.
Examples = TPCK for chymotrypsin; TLCK for trypsin
How is the Trojan Horse analogy applied in the concept of affinity labeling?
Enzyme active site = city of troy
Parts of inhibitor = trojans take into city as a victory trophy; the horse
Greek soldiers hidden inside represent the part of the inhibitor that resulted in destruction of city/inhibited enzyme
In the case of chymotrypsin and trypsin, what is the war head?
wants to react with histidine
chloromethyl ketone group
reactive part that reacts with critical active site of histidine residue
What’s the significance of TPCK for chymotrypsin and TLCK for trypsin?
TPCK doesn’t react with trypsin
TLCK doesn’t react with chymotrypsin
Difficult to freely isolate chymotrypsin and trypsin from each other; always stuck with contamination
you can purchase trypsin that has been TPCK treated
means that activity has been knocked out and leaves trypsin alone (same deal with TLCK + chymotrypsin)