Enhancer sequences + Activator/ Repressor proteins
1/16
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
17 Terms
What are features of enhancer sequences?
Additional Cis-acting element (like the promoter) required for maximal gene transcription
Can be upstream / downstream from transcription initiation site & can modulate despite being thousands of bp away.
Have short sequence elements like promoter seq, which activators can bind to forming protein-complexes → allows enhancer complex to be close to the promoter, increasing transcription
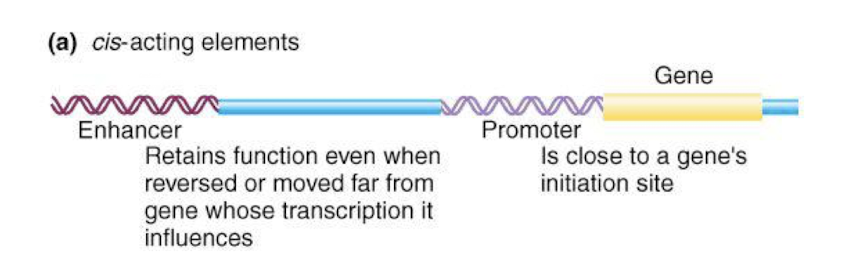
How do Enhancers work if they are far away from the promoter?
Enhancers can loop through the DNA to bring the bound activators in proximity to the promoter.
This looping facilitates the recruitment of additional transcription factors and RNA polymerase, enhancing transcription despite the distance.
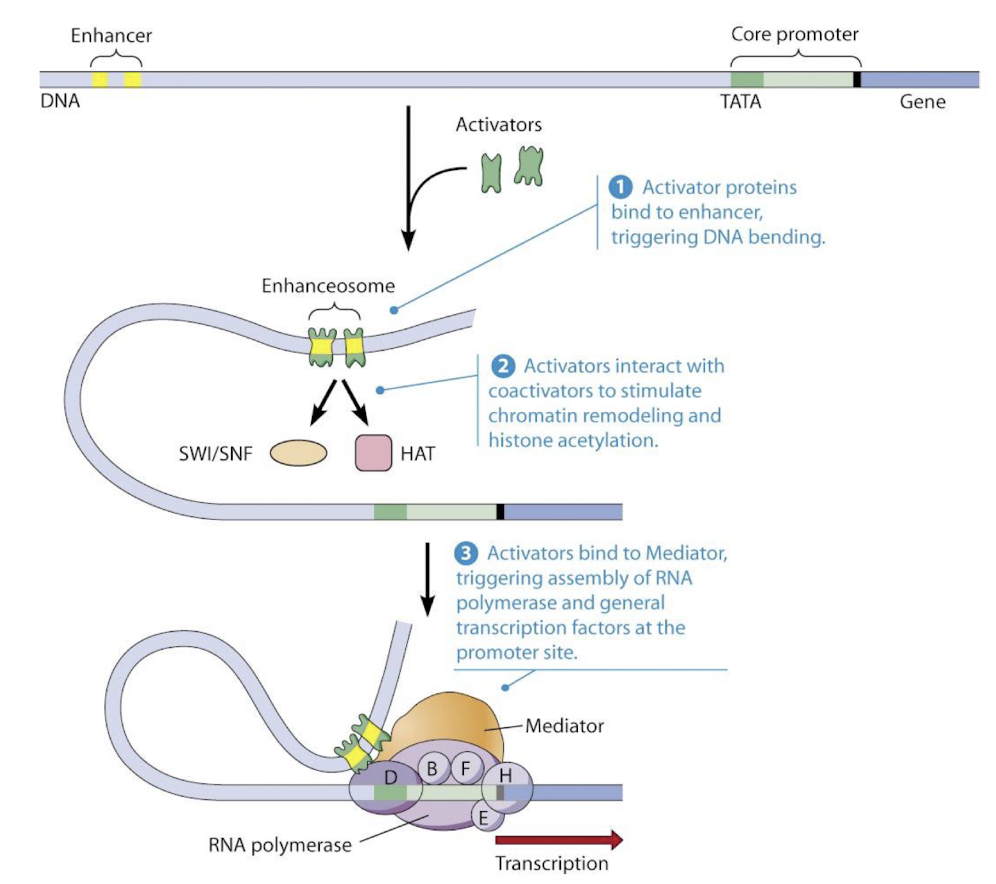
Structure of a TF review
Comprised of a:
DNA-binding domain - interacting with specific DNA sequences
Activation domain - interacts with other proteins to stimulaate transcription from a nearby promoter
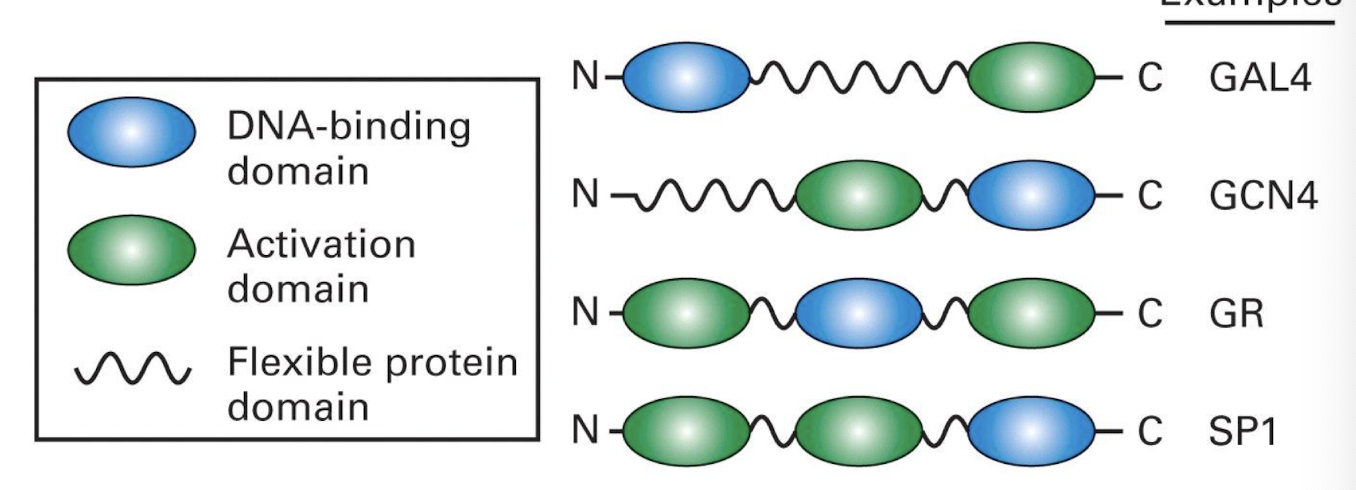
What is the difference between enhancers / silencers?
Specific Transcription factors bind to specific sequences some distance from the core promoter, named enhancers (positive effect) or silencers (negative effect)
What are some structural properties of enhancers?
Range in length from about 50 to 200 base pairs and include binding sites for several transcription factors
Multiple transcription factors bind to a single enhancer and interact
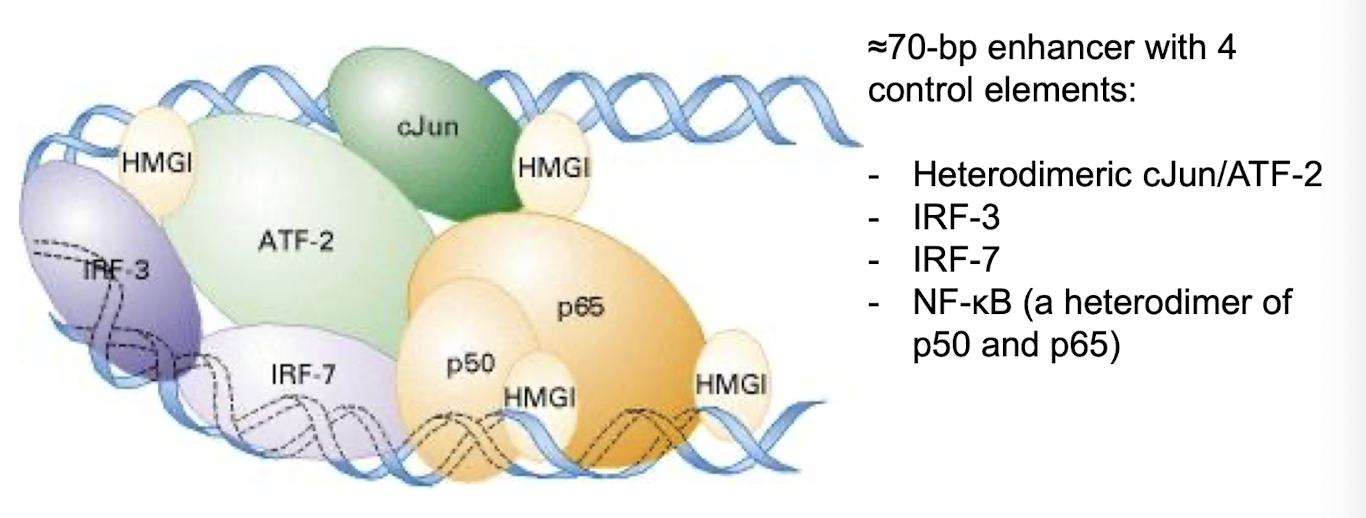
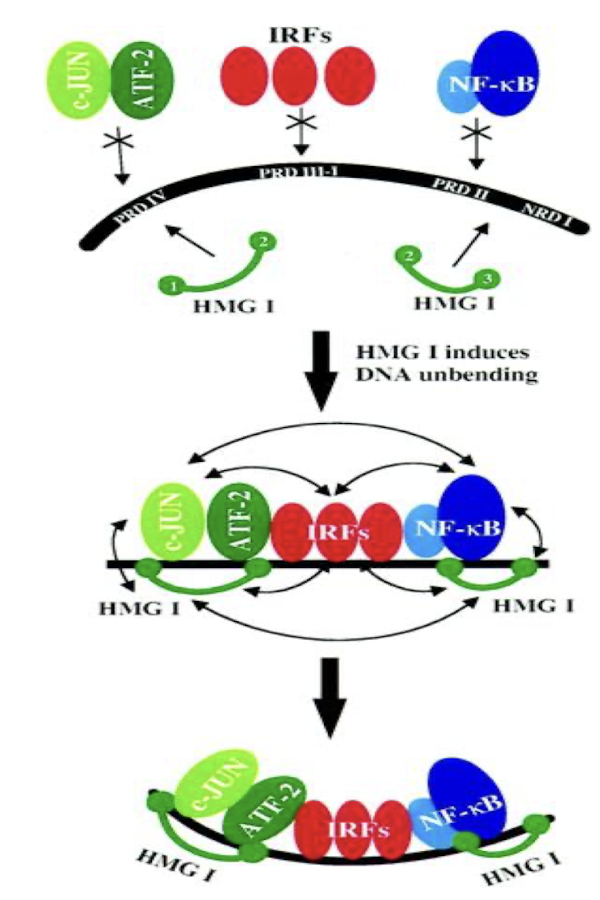
What is an example of a multiprotein complex that binds to enhancer sequences?
β-interferon, an important protein in defence against viral infections in humans.
• Expression controlled at β-interferon 70-bp enhancer comprised of four control elements
What are the 4 control elements of the 70-bp enhancer - β-interferon enhancesome?
Heterodimeric cJun/ATF-2
IRF-3
IRF-7
NF-kB - heterodimer of p50 and p65
What are the 2 steps of the enhanceosome assembly?
HMG1 binds to the minor groove causing unbending of DNA enhances protein-protein interaction for next step, leads to stable nucleoprotein structure
TF binding
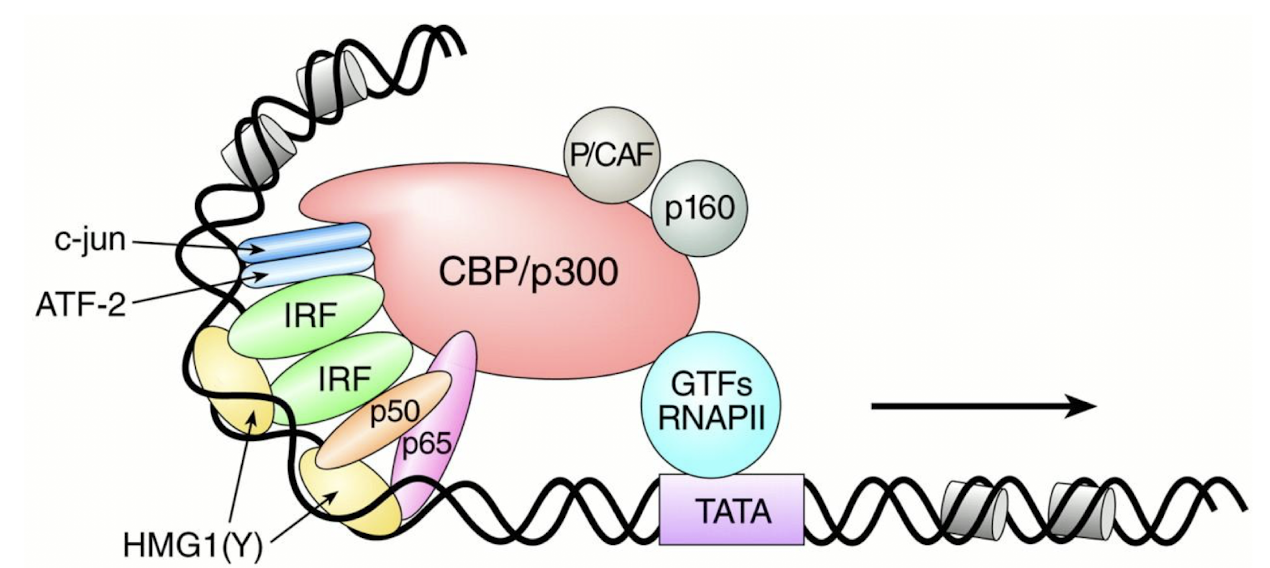
What do co-activators do and what is an example?
Co-activators are proteins that increase transcription of specific genes by interacting with activators and the basal transcription machinery.
An example is the CBP/p300 protein - recruited by the β-IFN enhanceosome, which enhances gene expression by serving as a bridge between GTF and RNA polymerase.
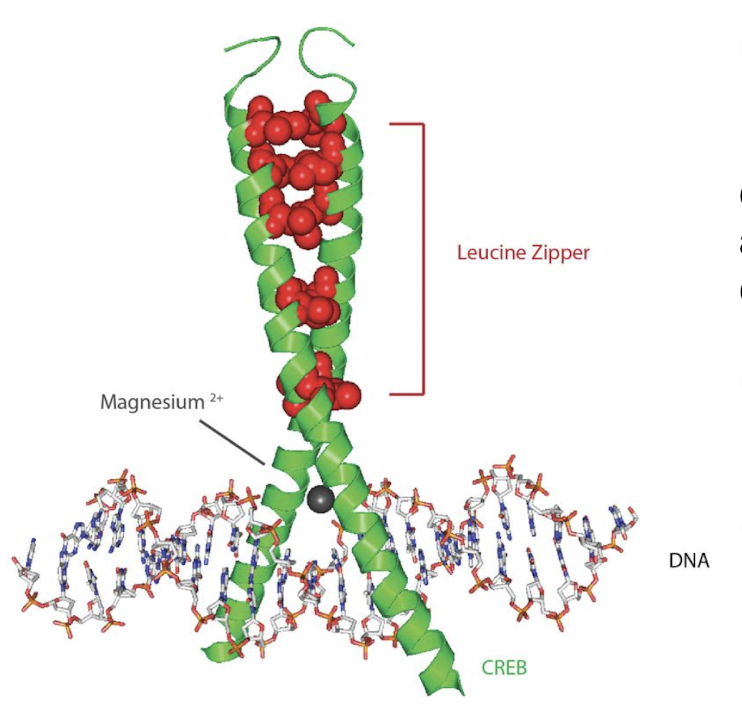
What is another example of a TF that acts as a co-activator?
CREB - cAMP response element binding protein
Binds to CRE on target gene (cAMP response element)
What are some features of the CREB cellular TF?
It is a Leu zipper structure that facilitates dimerization by its 2 helical coils
Regulates transcription of genes such as neuropeptides e.g. somatostatin and genes involved in the mammalian circadian clock (PER1, PER2)
Functions in neuronal plasticity, long term memory + loss of function is linked with Alzheimer’s
CREB is also phosphorylated by protein kinase A (PKA)
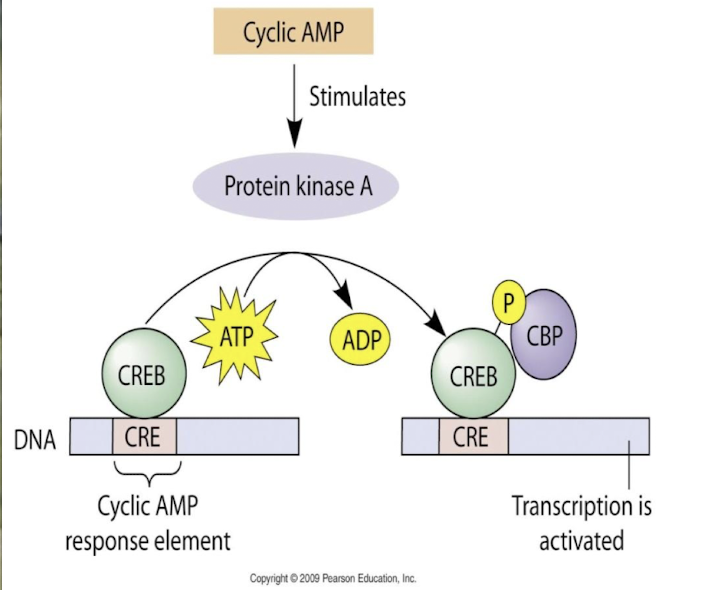
How does the activation domain of CREB initiate transcription?
CREB AD binds to co-activator CBP (CREB-binding protein)
PKA phosphorylates Ser on CREB
CREP~P doesn’t alter DNA binding properties, but binds to the co-activator CBP
This interaction recruits additional transcription machinery, i.e. GTF & RNAP II enhancing the transcription of target genes.
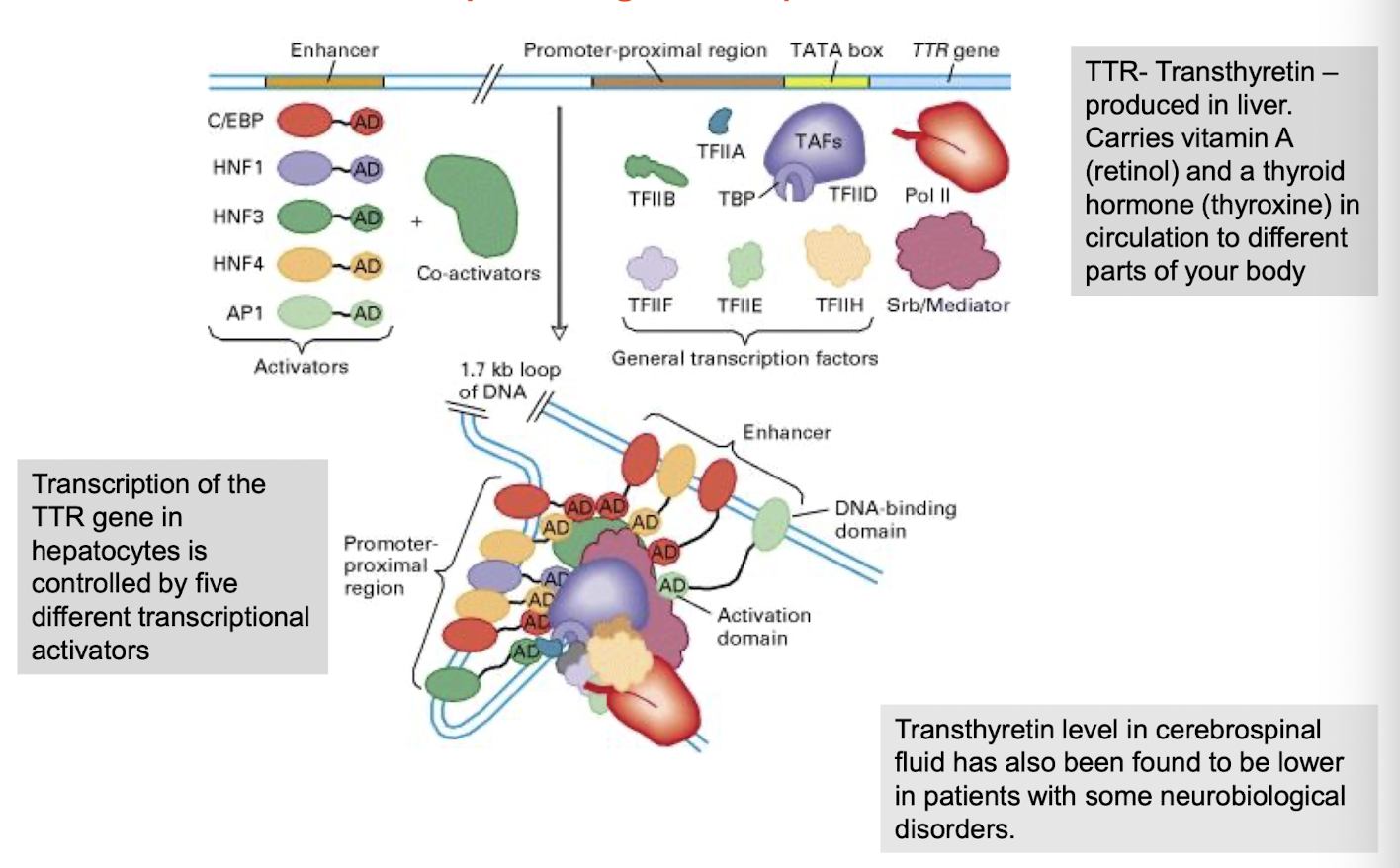
What is an example of cell-type specific gene expression?
Co-operative assembly of an activated transcription-initiation complex at the TTR promoter in hepatocytes
TTR = Transthyretin produced in liver + carries Vit A (retinol) & thyroxine in circulation to different parts of the body
How is TTR transcription controlled?
By 5 different transcriptional activators such as HNF1, HNF4, and C/EBP, which interact with enhancer elements in the TTR promoter region.
What are features of transcriptional repressors?
Act oppositely to transcriptional activators:
Mutation of an activator-binding site leads to decreased expression
Mutation of a repressor-binding site leads to increased expression.
• Like activators, repressors have two functional domains: a DNA-binding domain and a repression domain.
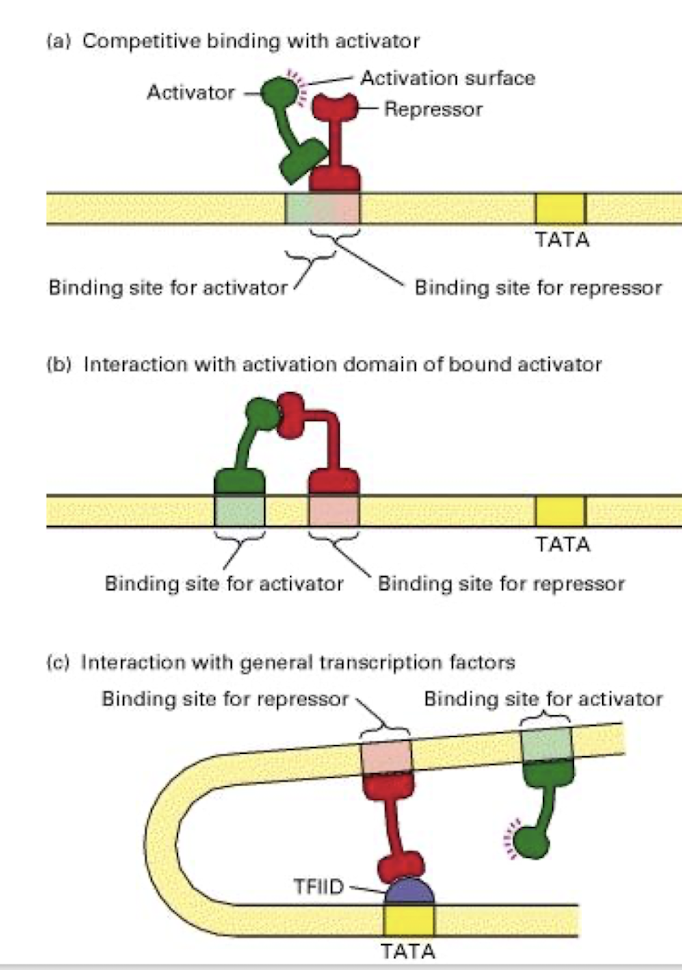
3 mechanisms of transcriptional repression
Competitive binding with activator for binding sites
Interaction with AD of bound activator
Interaction with GTF to prevent initiation
What is an example of how lack of appropriate repressor activity can cause disease?
Wilms tumour:
WT1 gene encodes a repressor protein that has a C2H2 ZF-DNA binding domain which binds to the control region of the EGR-1 transcriptional activator
Binding of WT1 represses EGR-1 gene
EGR-1 is a transcription factor that, when not mutated, can lead to no functional WT1 protein produced & overproduction of EGR-1 → kidney tumour development