Unit Three - Chapters 15 and 16
1/116
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
117 Terms
Transcription
The process of copying the sequences of DNA into an RNA molecule, which occurs in the nucleus of a eukaryotic cell and is catalyzed by an RNA polymerase.
1) It uses RNA polymerase
2) The RNA polymerase doesn’t need a primer
3) The RNA product doesn’t last
4) It has no proofreading mechanisms, meaning it is less accurate
What four things about transcription are different than replication?
RNA Polymerase
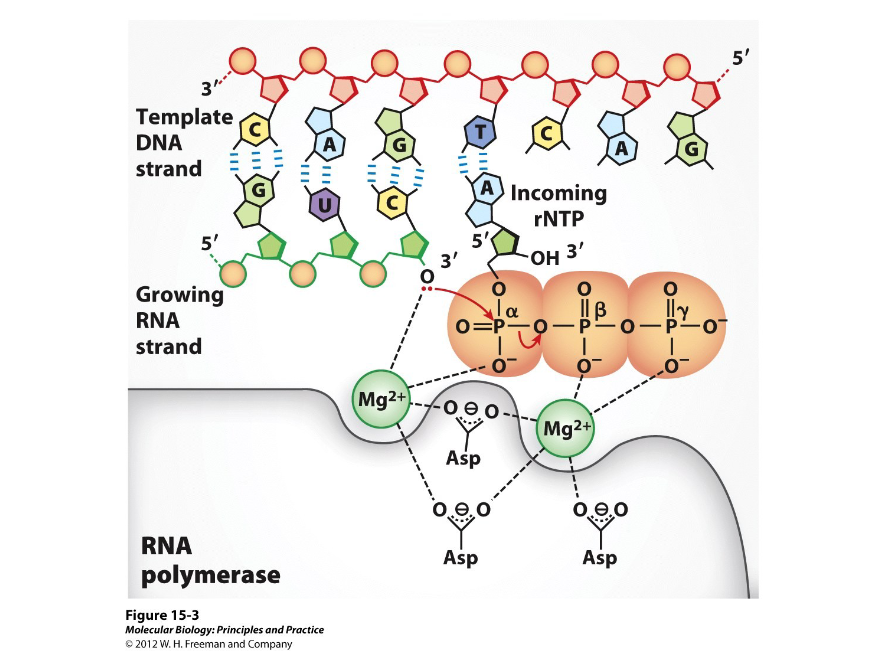
First discovered in E.coli in the 1960s, this is an enzyme that performs the same reaction in all cells and is known to require a DNA template. As with DNA synthesis, this enzyme synthesizes RNA 5’ to 3’ with the 3’ hydroxyl of the growing RNA strand attacking the incoming alpha phosphate.
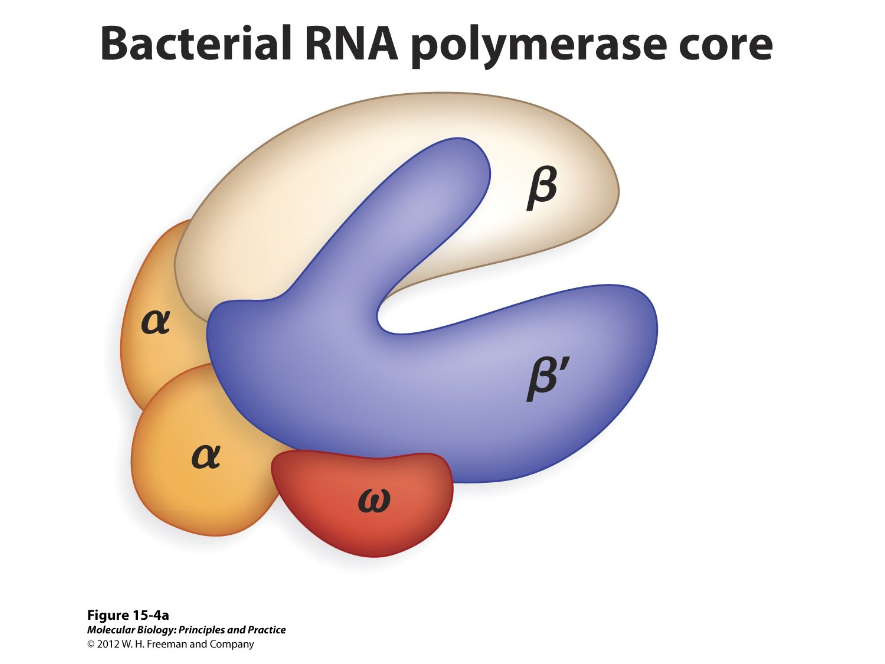
Structure
RNA polymerases vary in ( ); in some viruses, they consist of a single polypeptide, while in other cells, they consist of multiple subunits.
1) B (large)
2) B’ (large)
3) 2 alpha (small)
4) Phi (small)
In bacteria, the core of a RNA polymerase consists of ( ) polypeptide subunits, two large and three small, which are…
Sigma Factor
A sixth polypeptide that binds transiently to the core of the RNA polymerase and directs the enzyme to the promoter. This structure plus the core forms the RNA polymerase holoenzyme.
1) Initiation
2) Elongation
3) Termination
What are the three stages of transcription?
Promoters
DNA sequences that mark the beginning of a gene. Initiation begins at these defined sites.
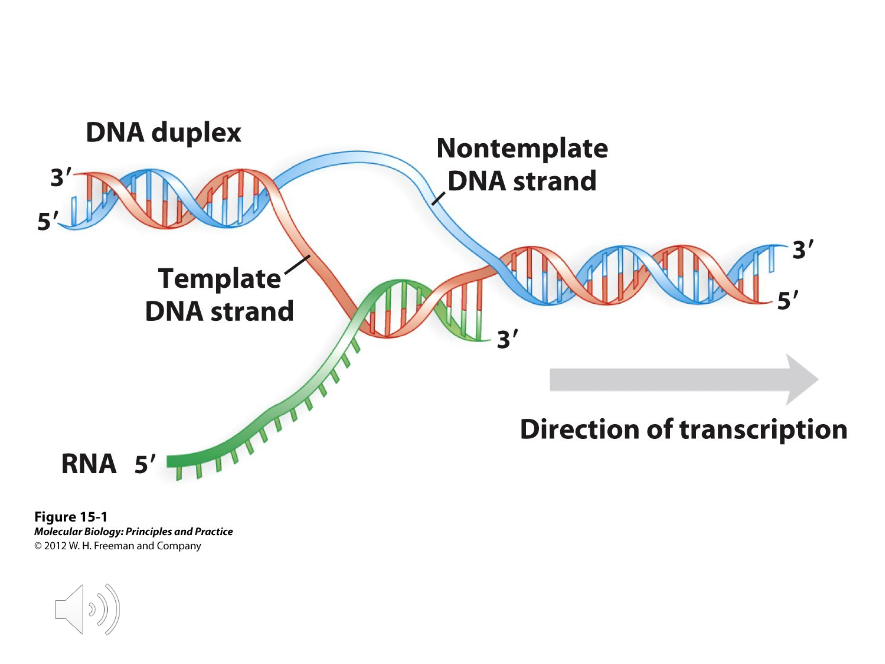
Template Strand
The DNA strand that serves as the pattern for the newly-synthesized RNA strand in Initiation of transcription.
Non-Template Strand
The DNA strand that is not used during Initiation of transcription but is identical to the newly-synthesized RNA strand in all regards except uracil and thymine.
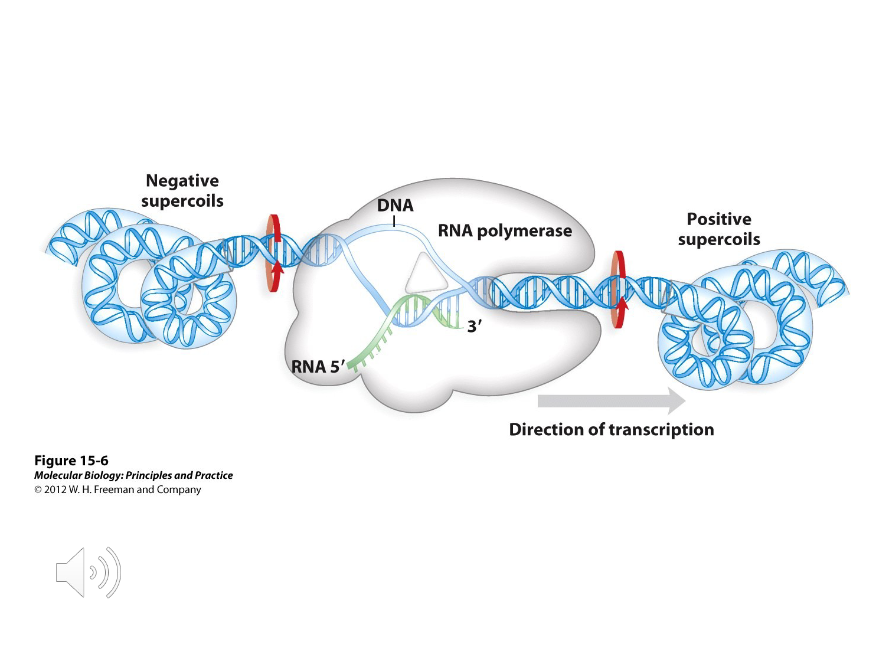
Transcription Bubble
The short distance that DNA is unwound for transcription. In bacteria, this is about 17 base pairs in length total, with the RNA/DNA hybrid taking up about 8 base pairs.
Positive/negative
(Positive/negative) supercoils will form at the front of the transcription bubble while (positive/negative) supercoils will forms at the back of the bubble.
1) Open Complex
2) Closed Complex
Initiation of transcription goes through two defined steps, which are…
Closed Promoter Complex
The phase of initiation of transciption that involves the binding of the RNA polymerase to the promoter. The enzyme is bound to one face of the helix and the DNA stays double stranded.
Open Promoter Complex
The phase of initiation of transcription where DNA is opened up over a distance of about fourteen base pairs and the first ten bases of the RNA strand are synthesized. This short transcript of RNA is typically released and initiation begins again.
This phase is also known as the initial transcribing complex.
Elongation
When an enzyme has added more than the 10 bases to the transcript, it is said to have escaped or cleared the promoter. This begins the process of ( ).
Elongation Complex
This complex is formed during elongation when RNA polymerase binds to the DNA and completes transcription of the RNA transcript.
Sigma 70
The main sigma factor in E.coli.
Six
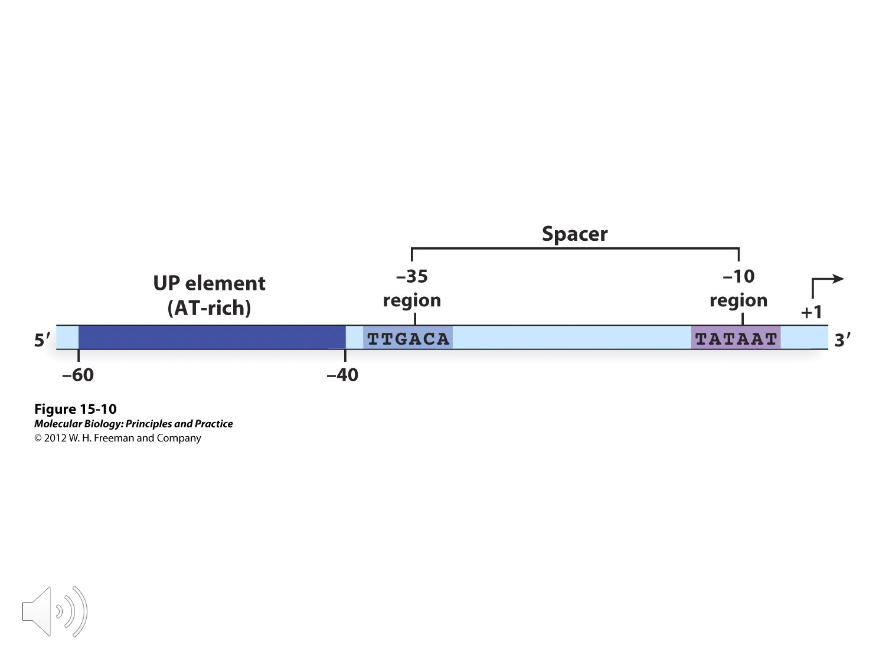
In E.coli, the promoters recognized by sigma 70 consist of two ( )-base sequences separated by eighteen bases. Doing the math, these sequences are located at the -10 and -35 regions of the E.coli promoter, but they aren’t identical in all promoters.
TATAAT
TTGACA
By analyzing the sequences of the -10 and -35 regions of different E.coli promoters, consensus sequences have been averaged and they are, respectively,…
UP Elements
Sequences rich in AT base pairs that occur between -40 and -60 in especially strong promoters.
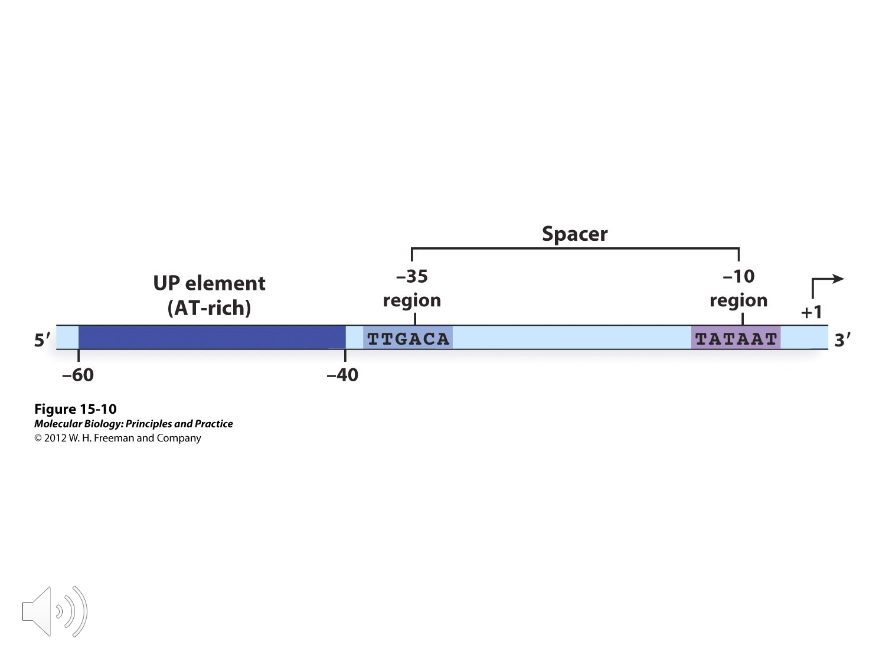
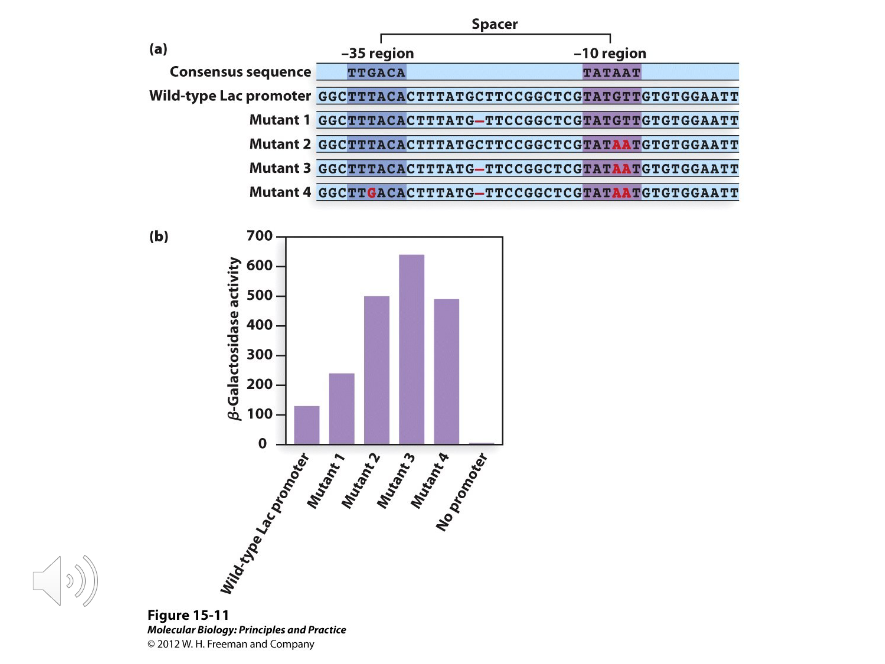
Promoter Up Mutations
Mutations that made the promoter more efficient.
Promoter Down Mutations
Mutations that made the promoter less efficient.
Two
The consensus sequences of the -10 and -35 regions were based on probabilities defined by ( ) types of mutations; promoter-up and promoter-down mutations.
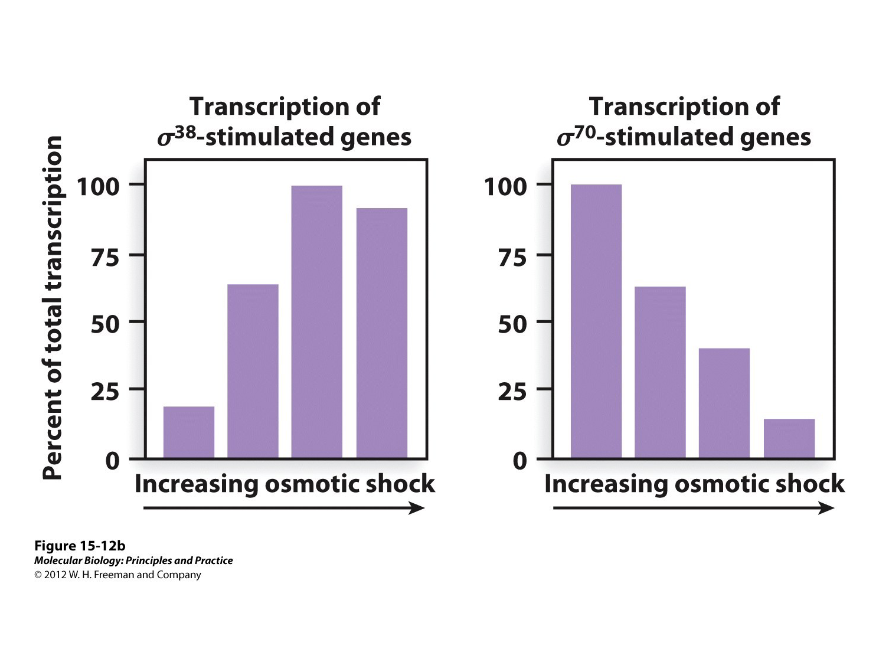
Sigma Factors
Bacteria have just one RNA polymerase, but they can have multiple ( ) recognizing different promoters.
Left matches starvation/stationary phase sigma factor
Right matches housekeeping sigma factor
This graph compares the efficiency of transcription of certain genes with increasing osmotic shock. If one sigma factor controlled genes that dealt with starvation and the stationary growth phase while the other controlled housekeeping genes, which graph would match the transcriptional activity of the enzyme during osmotic shock?
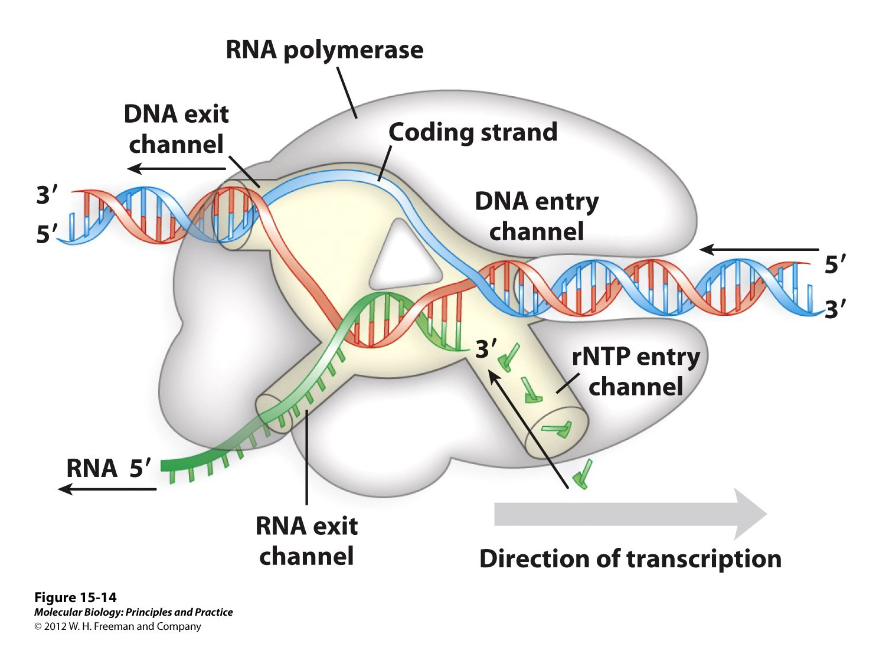
1) DNA exit channel
2) DNA entry channel
3) rNTP entry channel
4) RNA exit channel
What are the four different channels on a RNA polymerase?
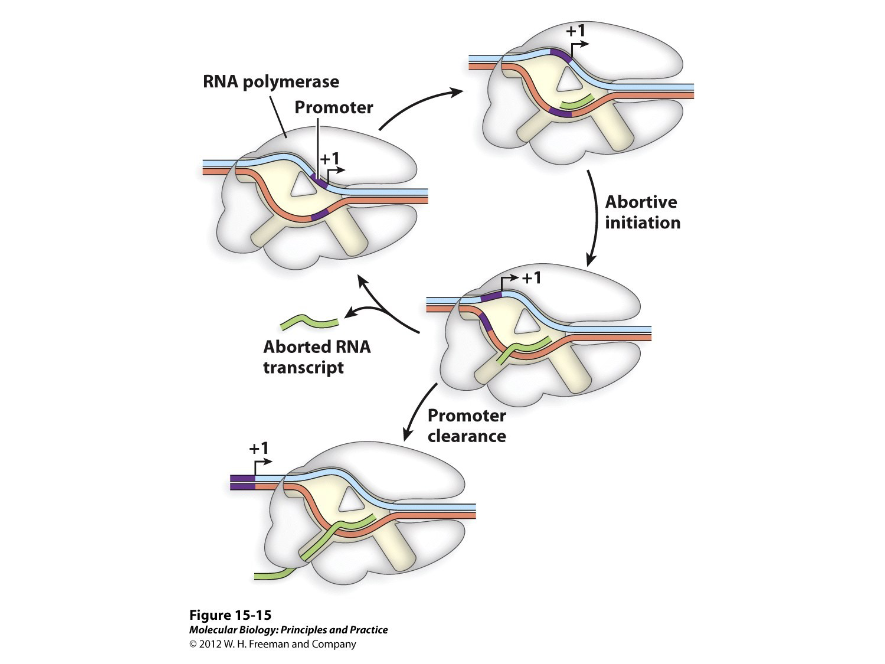
Abortive Initiation
This occurs during the synthesis of the first ten bases of a RNA transcript. To synthesize, the RNA polymerase has to collect and combine two rNTPs in the right orientation to move forward. As the first ten are synthesized, there is a high probability that transcription will abort and start over.
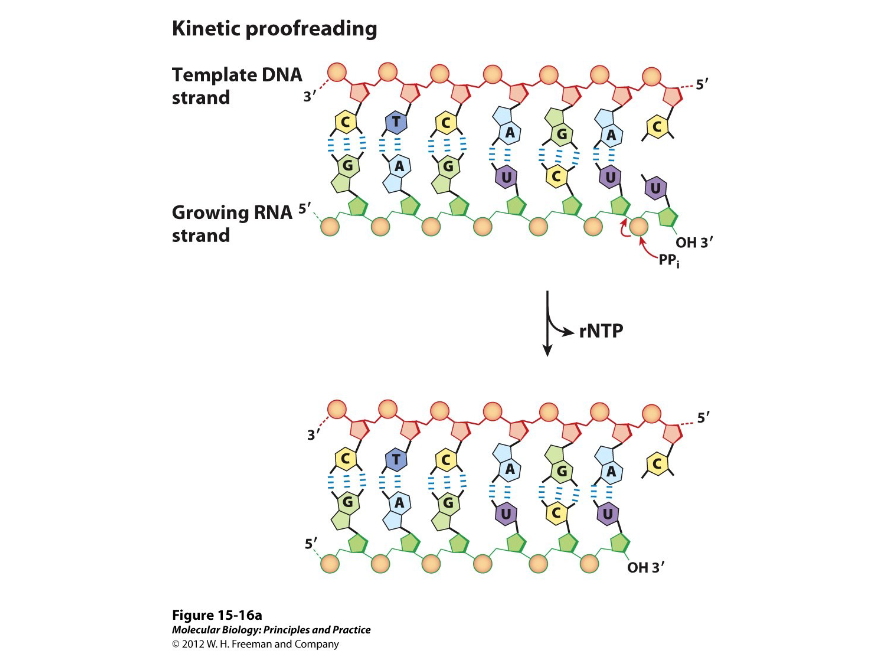
Pyrophosphorolytic Editing
Also called kinetic proofreading, this involves the enzyme using its active site to remove incorrect bases by reincorporating a pyrophosphate. The enzyme will briefly pause over the incorrect bases and replace them.
This occurs during elongation of transcription.
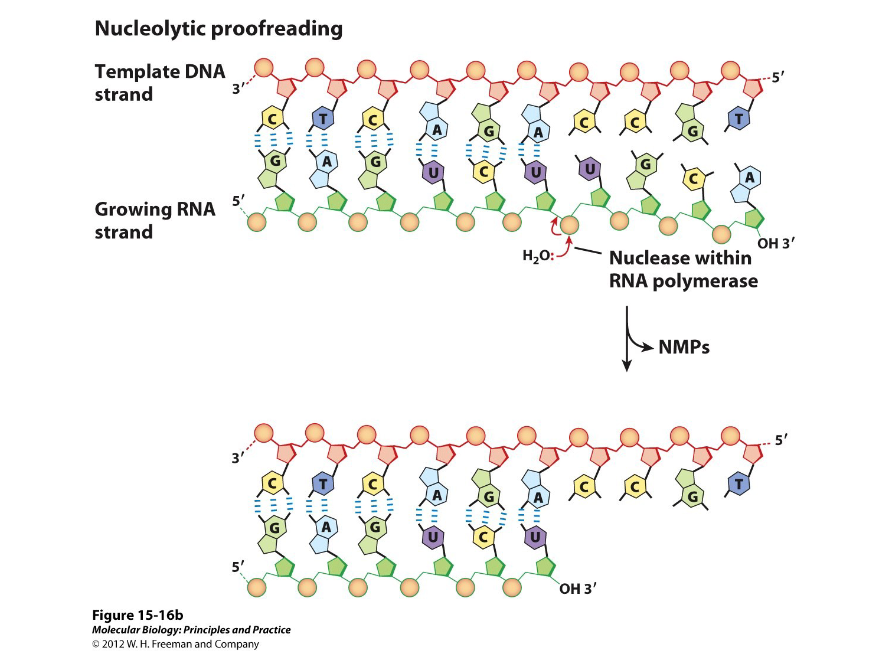
Hydrolytic Editing
During elongation of transcription, the enzyme will back up after a mistake, cleave the RNA product, and start the process of elongation over.
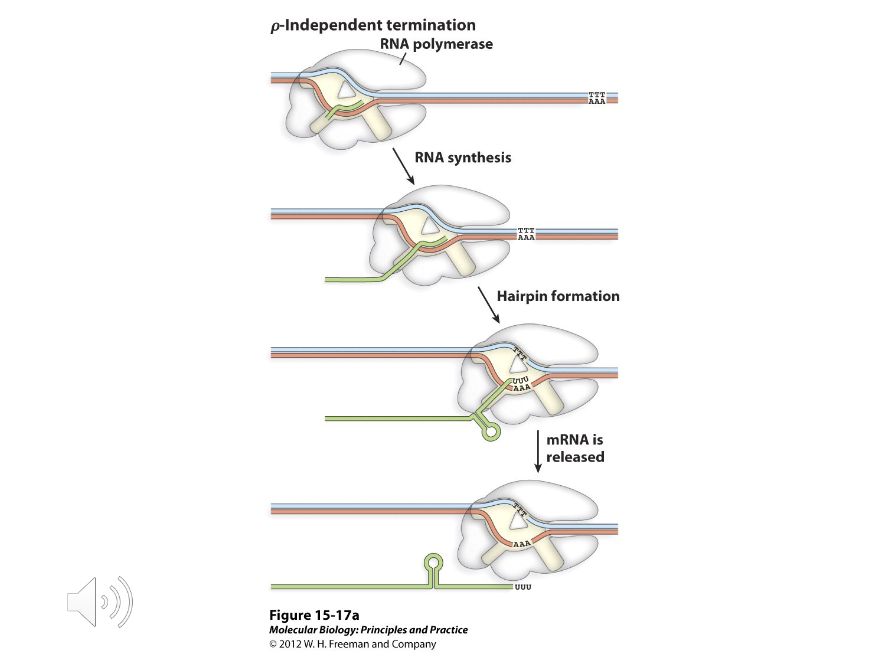
1) Rho independent
2) Rho dependent
E.coli has two classes of termination sequences, which are…
Rho Independent Terminators
The most common type of terminators that work solely on the basis on their shape. They are composed of a GC-base pair rich stem loop followed by a line of uracils.
Rho Dependent Terminators
Terminators that need a protein factor called Rho to work. They may consist of a stem loop that slows polymerase down while the Rho protein catches it and removes RNA from the template strand and RNA polymerase.
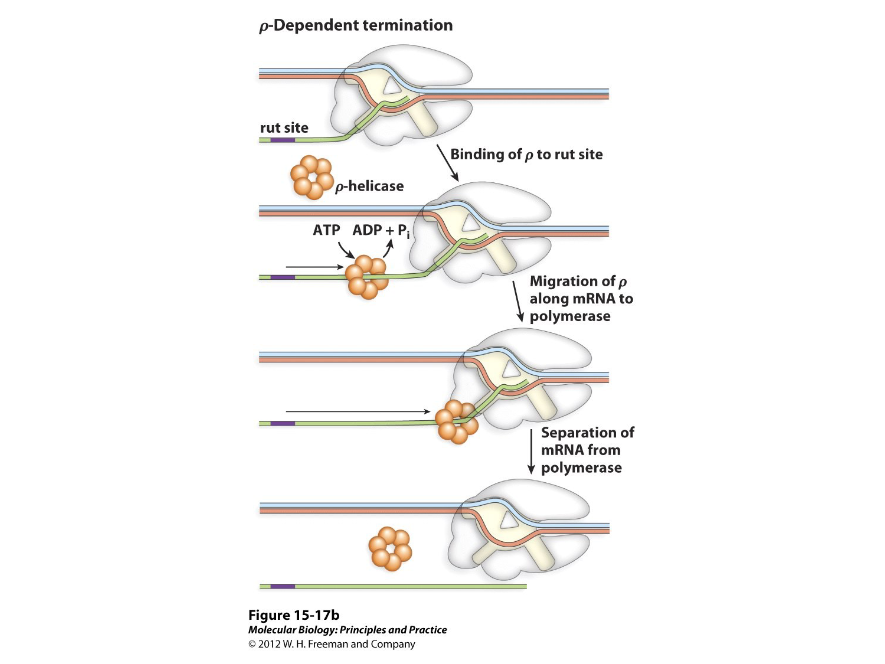
Three
Eukaryotes have ( ) different RNA polymerases.
Specific Transcription Factors
Transcription factors that bind DNA at long distances from various genes.
General Transcription Factors
Transcription factors that bind to the promoters of all genes.
Class/promoter
Each of the three RNA polymerases in eukaryotes only transcribe a certain ( ) of genes, have a specific type of ( ), and have their own transcription factors.
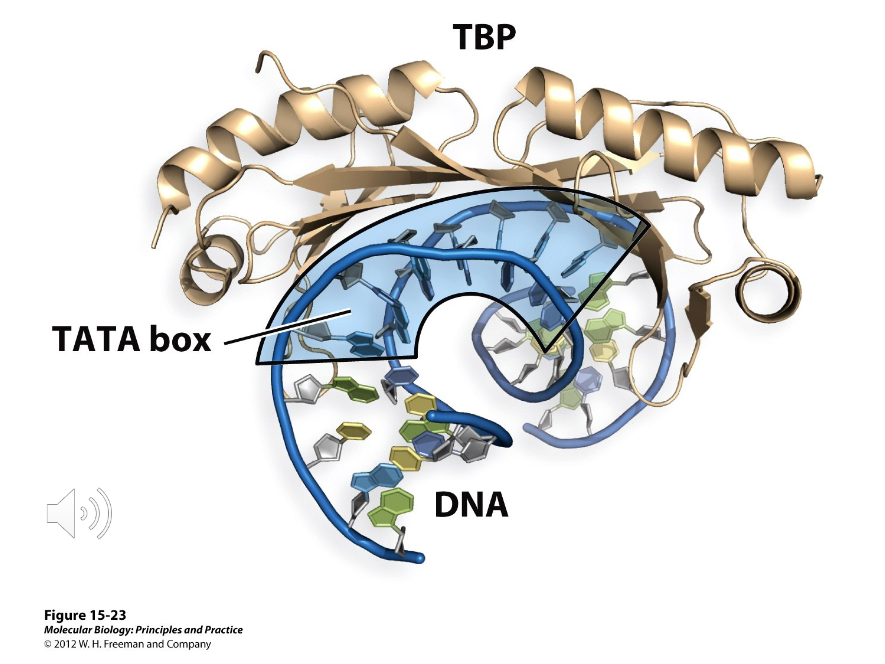
TATA-Binding Protein
A protein required by each polymerase in eukaryotes as part of their machinery. It binds to a sequence called the TATA box, but it is only present 25% of the time. It is highly conserved.
It works by contacting the minor groove of the DNA and distorting/bending the double helix.
RNA Polymerase I
One of the three eukaryotic RNA polymerases that transcribes only one gene, which encodes the precursor to ribosomal RNAs. There are many copies to this gene.
Start
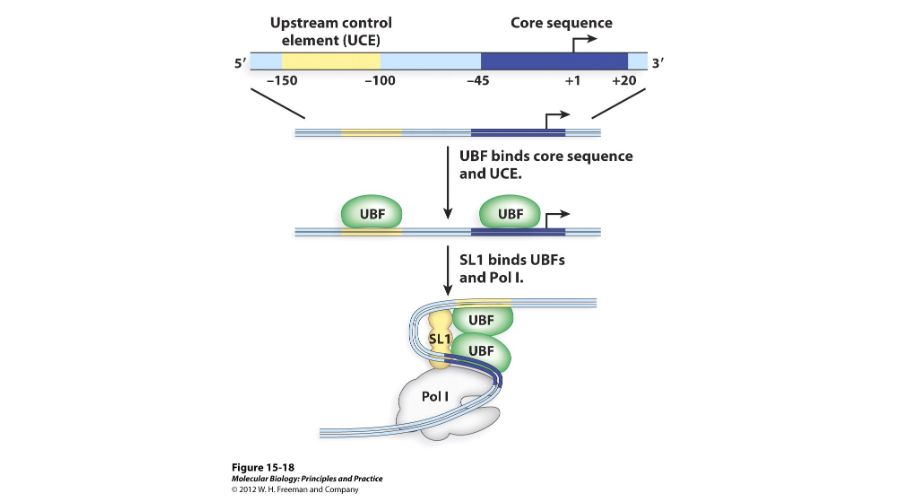
RNA polymerase I promoters have a core element and an upstream control element. The core element is located near the ( ) site of transcription, while the upstream control element is located at about -150 and -100.
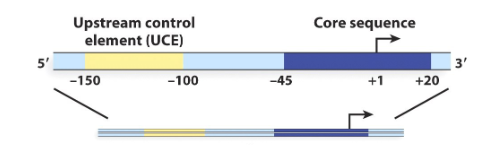
Two
RNA polymerase I has ( ) transcription factors; UBF and SL1. UBF, or the upstream binding factor, binds to the upstream control element and core element and recruits SL1. SL1 then binds near the core element.
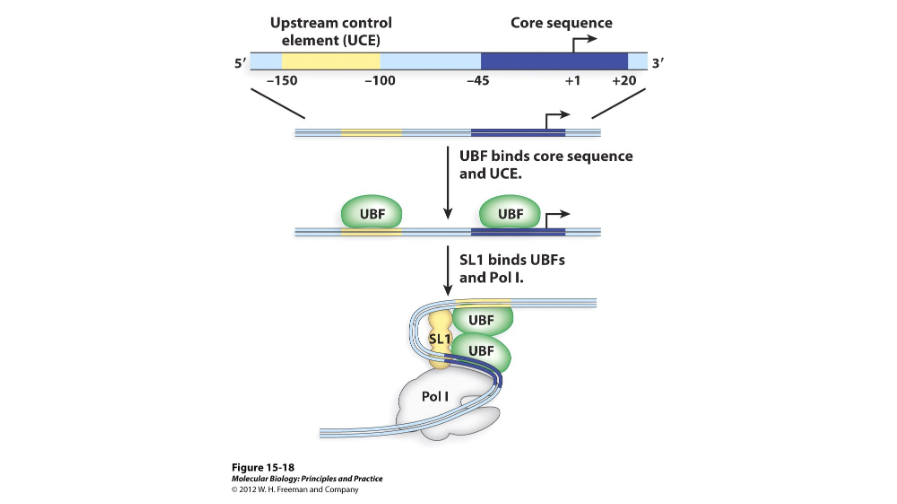
TBP/TAF
SL1 contains the ( ), or transcription binding protein, and the three ( ) molecules, or TBP-associated factors, needed to recruit RNA polymerase I.
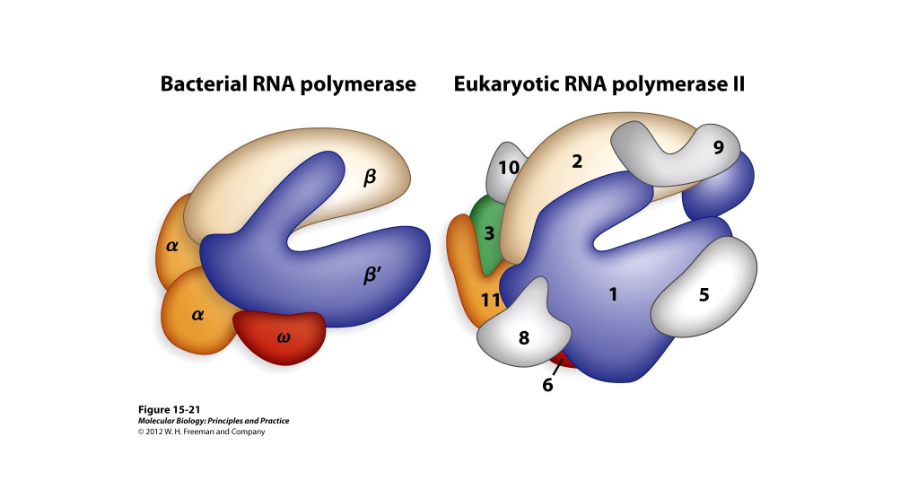
RNA Polymerase II
In eukaryotes, this makes mRNAs, or micro RNAs, from DNA transcripts. The eukaryotic version is more complex than the bacterial version but has structural similarities.
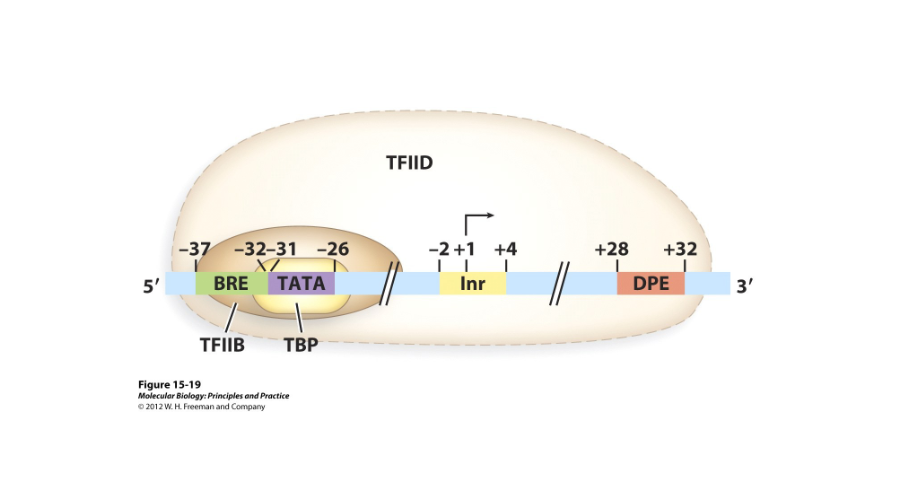
Start Site
The promoter for RNA polymerase II is usually about 40-60 bases long and can extend either upstream or downstream from the ( ). It contains the minimal set of sequences or elements required to activate transcription.
BRE
In the RNA polymerase II promoter, this is the recognition element for TFIIB, or the transcription factor for RNA polymerase II.
1) BRE, a TFIIB recognition element
2) A TATA element
3) An initiator (Inr)
4) Downstream promoter elements (DPE)
What are the core elements of a RNA polymerase II promoter?
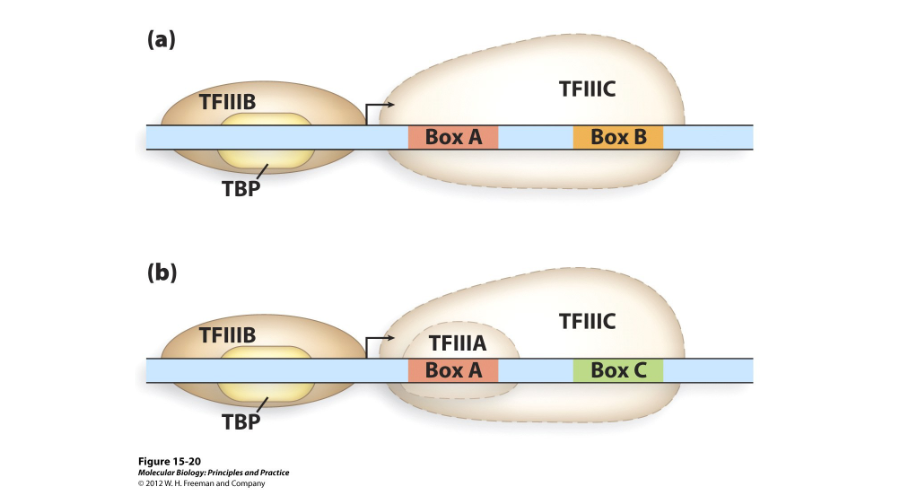
Thousands
Other sequences are needed for efficient transcription of most genes, but they are regulatory sequences that can be ( ) of base pairs away from the gene and are the target for specific transcription factors.
Downstream/two
Polymerase III promoters are located ( ) from the start site of transcription and contain ( ) sequence boxes. They require two transcription factors, though a third occasionally helps: TFIIIC, TFIIIB, and TFIIIA.
Pre-Initiation Complex
This contains RNA polymerase II and six general transcription factors.
1) TFIIA
2) TFIIB
3) TFIID
4) TFIIE
5) TFIIF
6) TFIIH
What are the general six transcription factors?
TFIID
A complex protein with many subunits that binds near the TATA box. It contains the TATA binding protein (TBP) and the TATA associated factors (TAFs).
Six
The TATA-binding protein associates with about ( ) TATA associated factors (TAFs) and binds to other promoter elements like the Inr and the DPE.
TFIIA
A transcription factor that doesn’t seem to always be essential. However, it may stabilize the binding between TFIID and the promoter.
TFIIB
A transcription factor that binds with DNA, with the BRE, and Downstream of the TATA box. It needs TFIID in order to bind.
TFIIF
A transcription factor that comes in with RNA polymerase and loosens non-specific attractions between DNA and polymerase II.
Closed promoter complex
TFIIE and TFIIH bind to create the ( ).
TFIIH
A transcription factor that has helicase activity and unwinds the DNA to form the open complex.
Promoter/released
At the end of elongation, RNA polymerase II is phosphorylated on its c-terminal domain (CTD) to force it to leave the ( ). Also, TFIIE and TFIIH are ( ).
Elongation Factors
These help the polymerase prevent pausing of transcription and enhance editing of the RNA transcripts.
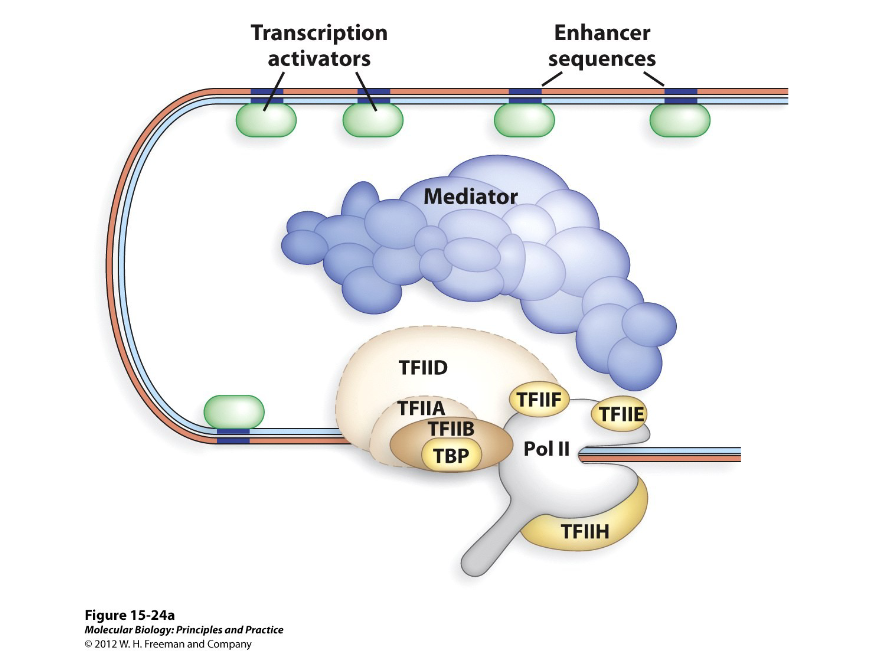
Mediator Complexes
This aids in the assembly of transcription factors around the promoter, meaning they are necessary for specific transcription factors to activate genes.
Anywhere
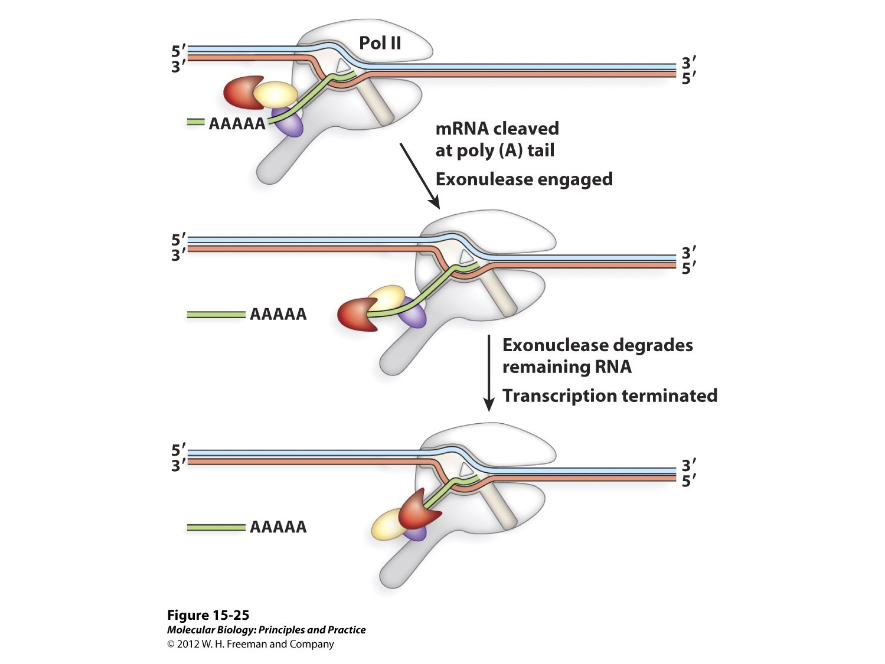
Polymerase II can terminate ( ), from a few base pairs away to several thousand base pair away from the end of the mature transcript.
Uncapped
In termination, an enzyme recognizes an ( ) RNA fragment left after cleavage and degrades the RNA from 5’ to 3’.
Primary Transcripts
RNA that is synthesized as biologically inactive precursors.
Aren’t
Most bacterial RNAs (are/aren’t) processed.
Capped/tailed/introns
Eukaryotic RNAs are ( ), ( ), and have their ( ) removed. Many of the enzymes involved in these modifications ride on the c-terminal domain of the RNA polymerase.
5’ Cap
An extra nucleotide, 7-methyl guanosine, linked by a 5’ to 5’ triphosphate linkage to RNA.
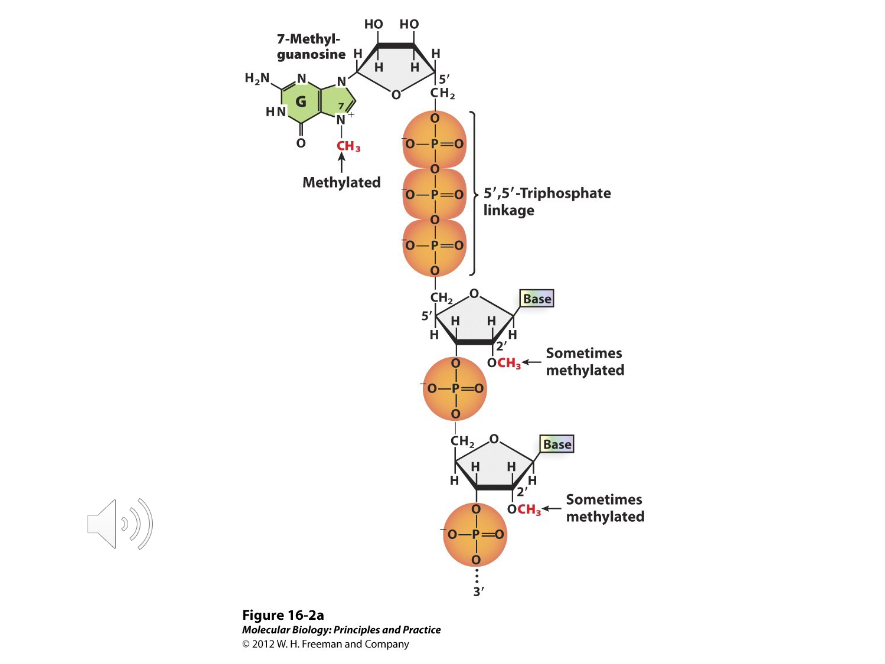
Phosphate
To synthesize a 5’ cap, the last ( ) of the 5’ PO4 on a new RNA is clipped off, GMP is added, and then methylated.
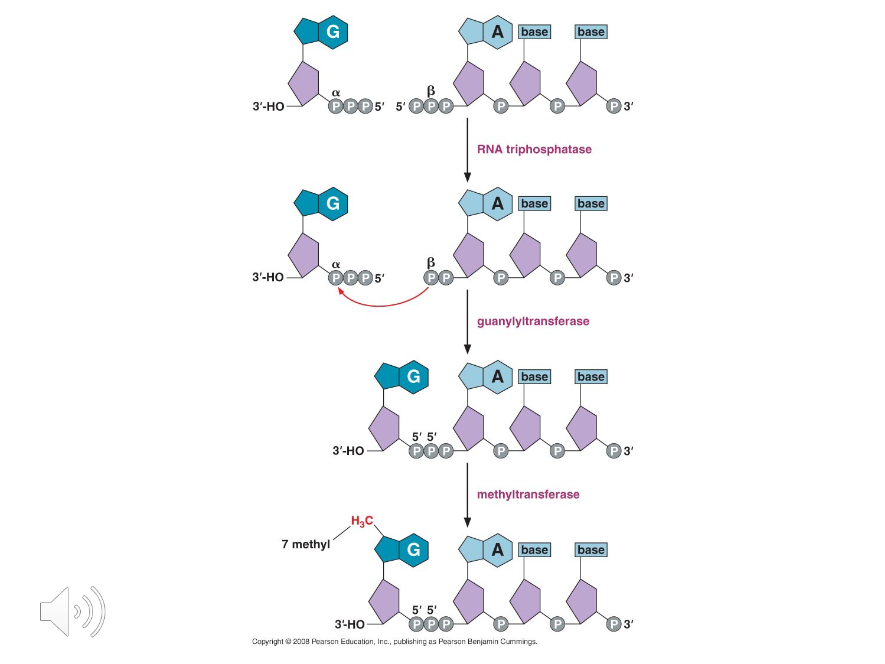
Guanylyltransferase
The capping enzyme.
mRNAs
Only ( ) are capped, so no capping enzymes associate with polymerase I or polymerase III.
Protects
Capping not only marks the mRNA for translation, but it also ( ) the mRNA.
Polyadenylation Tail
An RNA-modifier attached to the 3’ end of RNA. It is typically 80-250 bases long and protects mRNA from enzymatic destruction. Often, the c-terminal domain of the polymerase recruits poly-A enzymes that then attach this to the RNA.
After
Polyadenylation occurs ( ) the primary transcript is cleaved.
AAUAAA
In the pre-cleavage complex, what sequence of base pairs occurs about 10-30 base pairs before the poly A tail?

G/U
In the pre-cleavage complex, sequences rich in ( ) and ( ) lie 20-40 base pairs Downstream from the AAUAAA sequence.
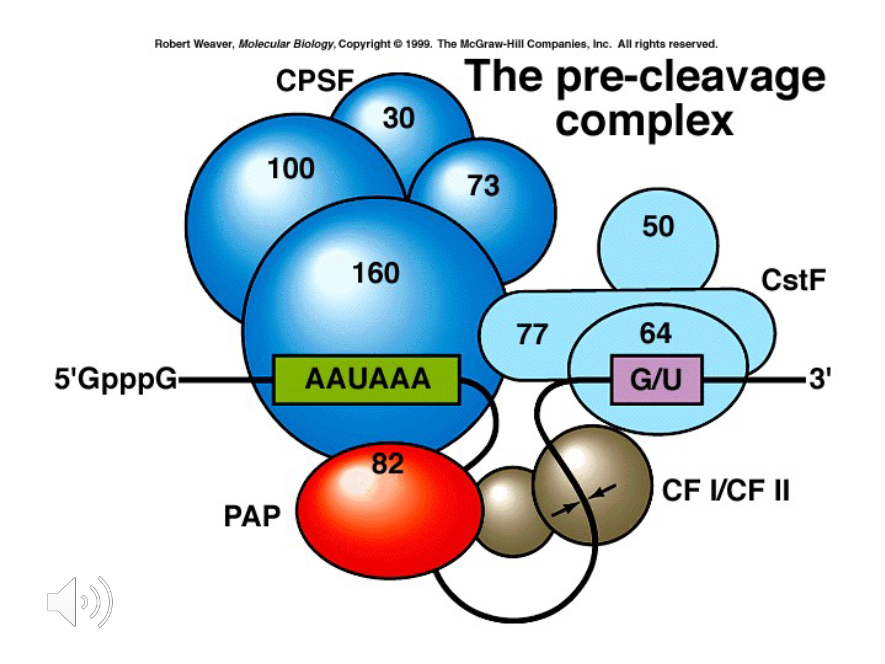
1) CPSF
2) CSTF
3) CF1 and CF2
What are the proteins involved in polyadenylation?
CPSF
A protein that binds to the AAUAAA signal during polyadenylation.
Poly-A polymerase
Once the polyadenylation factors cleave the mRNA, the poly-A is synthesized by ( ) and attached to the RNA with PABP.
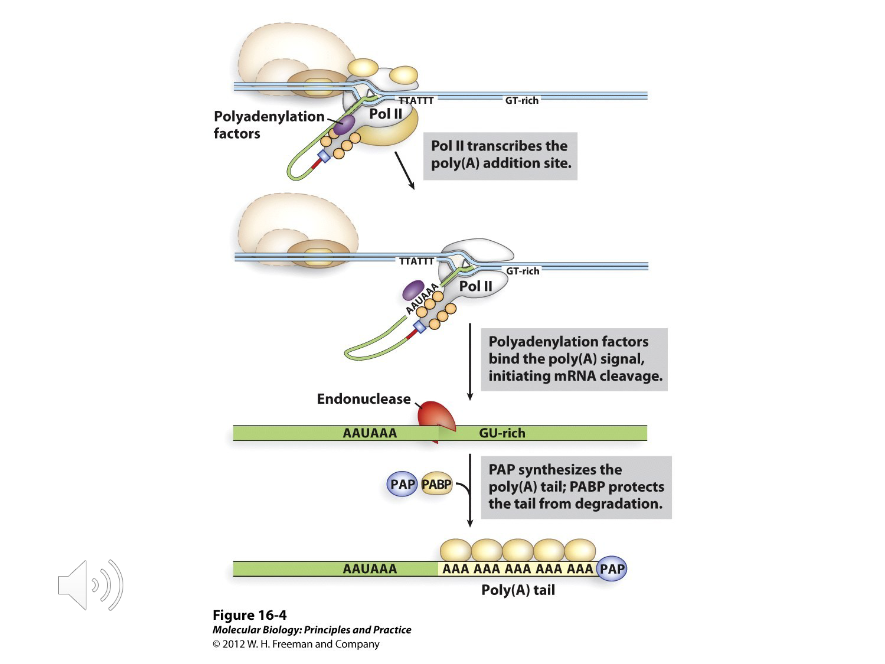
RNA Splicing
Further processing of RNAs which involves the removal of introns.
Contiguous
While genes are ( ) in prokaryotes, genes in eukaryotes are interrupted by large stretches of non-coding sequences.
Exons
The coding sequences of RNA.
Introns
Interruptions in genes.
Leder
Identified introns on the B-globin gene. He did this by separating double-stranded DNA into single strands and mixing them with mature mRNA of the gene. If the mRNA binds to a region of a gene with one intron, two single stranded DNA loops will form that are separated by a double-stranded DNA region.

Alternative Splicing
( ) increases the number of polypeptides that can be produced from a single gene.
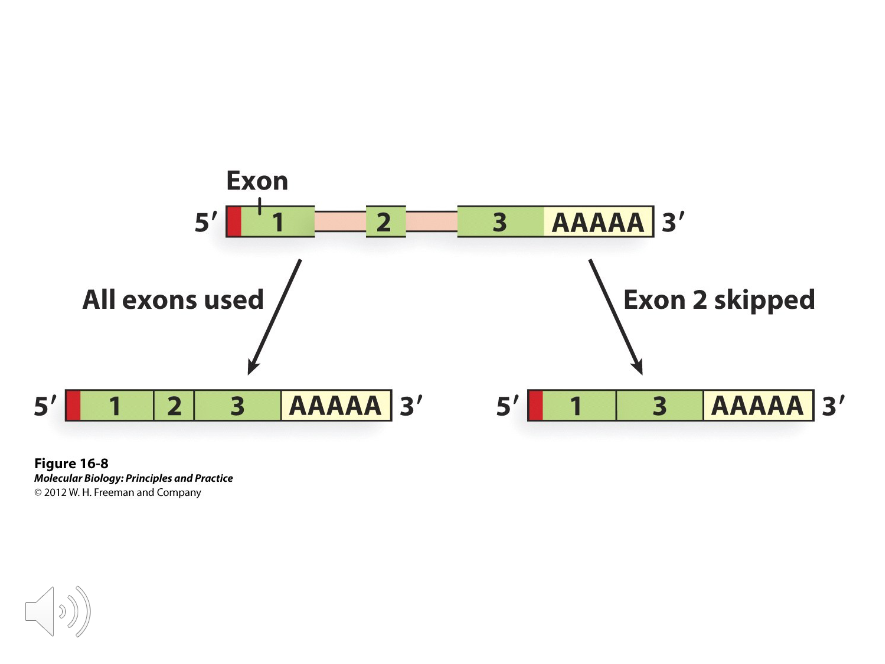
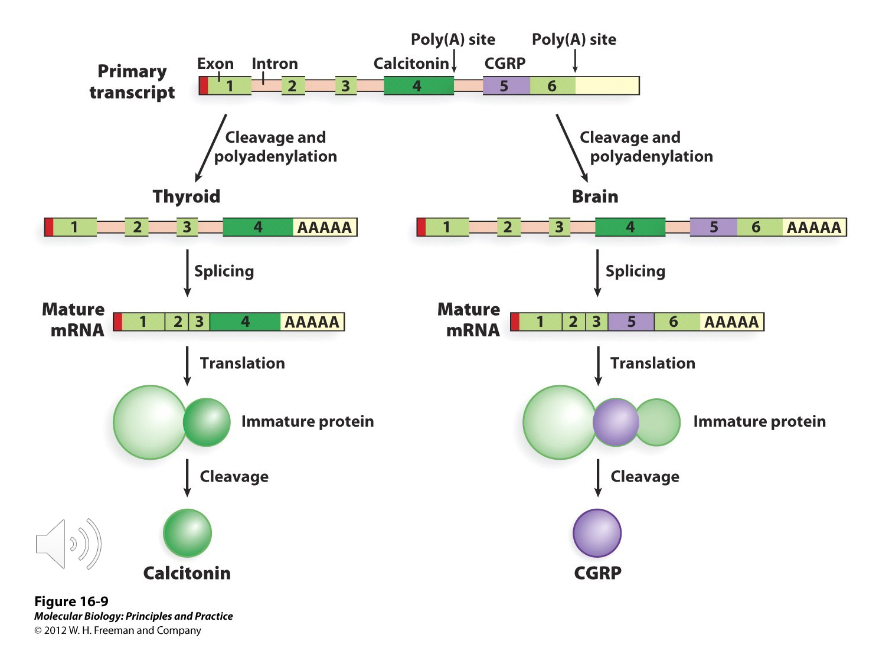
Poly-A
Using different ( ) sites in addition to alternative splicing of introns can also be used to create different gene products.
5’/3’/3’
The 5’ splice site marks the ( ) end of the intron and the 3’ splice site marks the ( ) end of the intron. The branch point site is found close to the ( ) end.
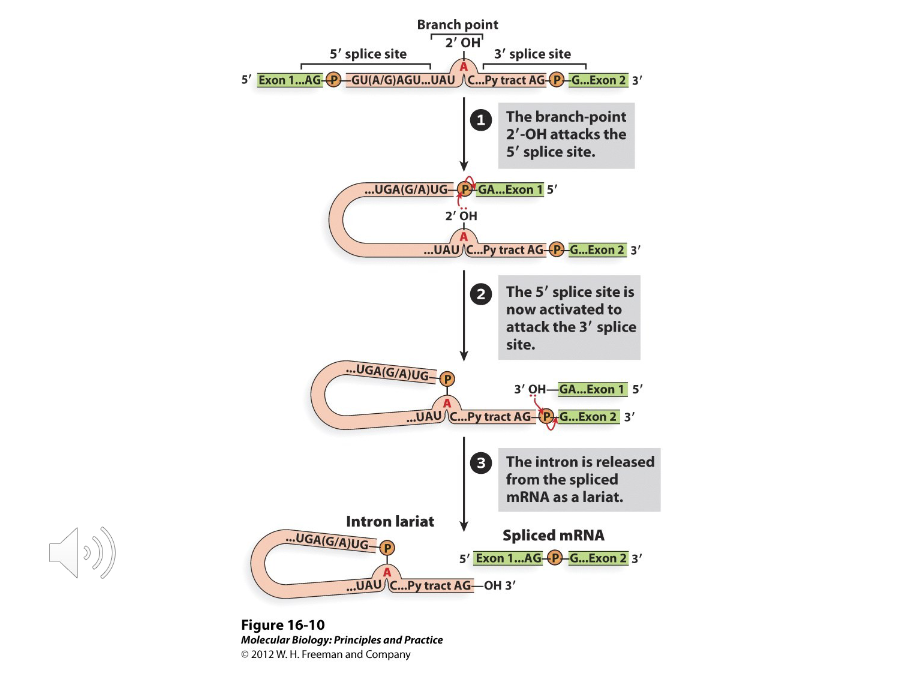
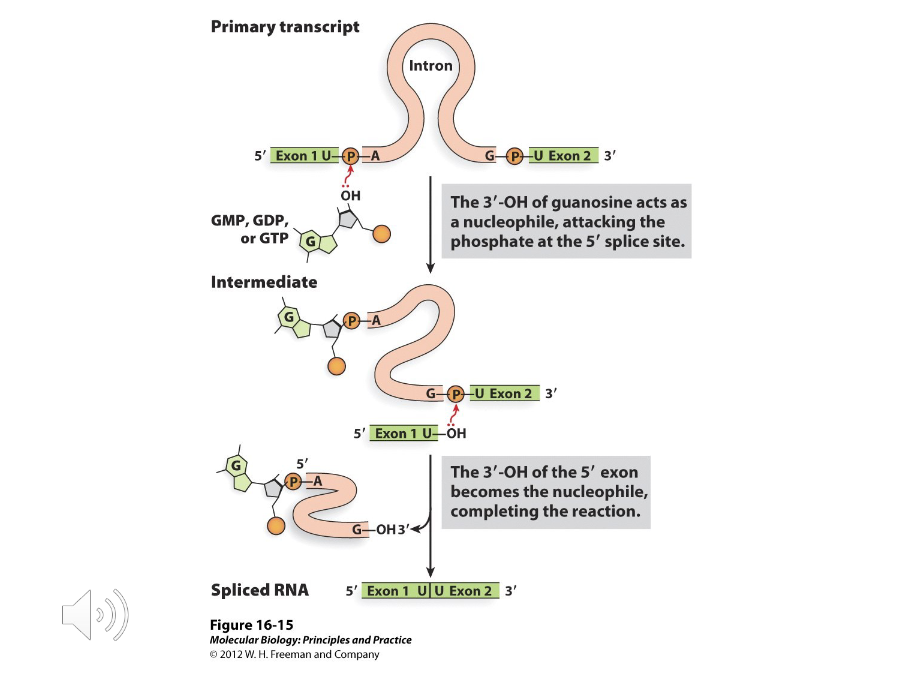
Lariat Model of Splicing
A model of splicing with two steps that includes phosphodiesterase exchange reactions. In these reactions, phosphodiester bonds in the primary transcript are broken and new ones are made. It is used by most nuclear genes.
1) The 2’ OH group of adenosine attack the phosphodiester bond between the first exon and the guanine at the beginning of the intron
2) The 3’ OH group at the left end of the first exon attacks the phosphodiester bond between guanine and the end base of the second exon
What are the steps of the Lariat model of splicing?
Splicosomes
Proteins that mediate the reactions of nuclear splicing. They contain 5 small nuclear ribonucleoproteins, also called SNRNPs (U1, U2, U4, U5, and U6), as well as hundreds of protein components.
snRNPs
Components of splicosomes that contain small nuclear RNA about 100 to 200 bases long.
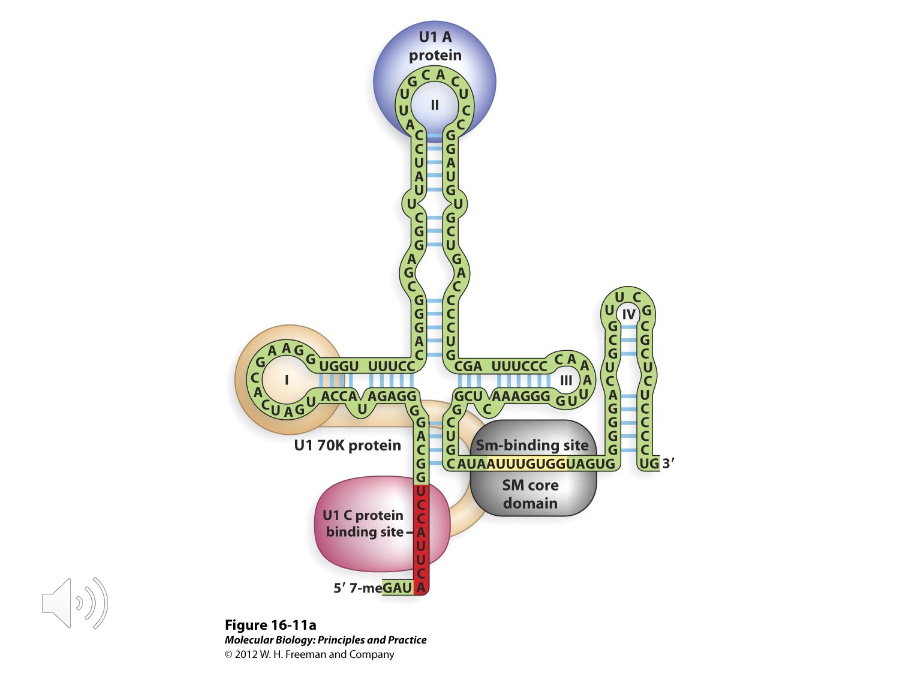
Sm Proteins
Each snRNA in a snRNP binds to a set of proteins called the ( ). They are common to all snRNPs and form the Sm core domain.
Sm Proteins
These proteins play important roles in selecting splice sites. They are part of the spliceosome but not a part of the snRNP.
GU/AG
Introns have the sequence ( ) on the 5’ end and ( ) on the 3’
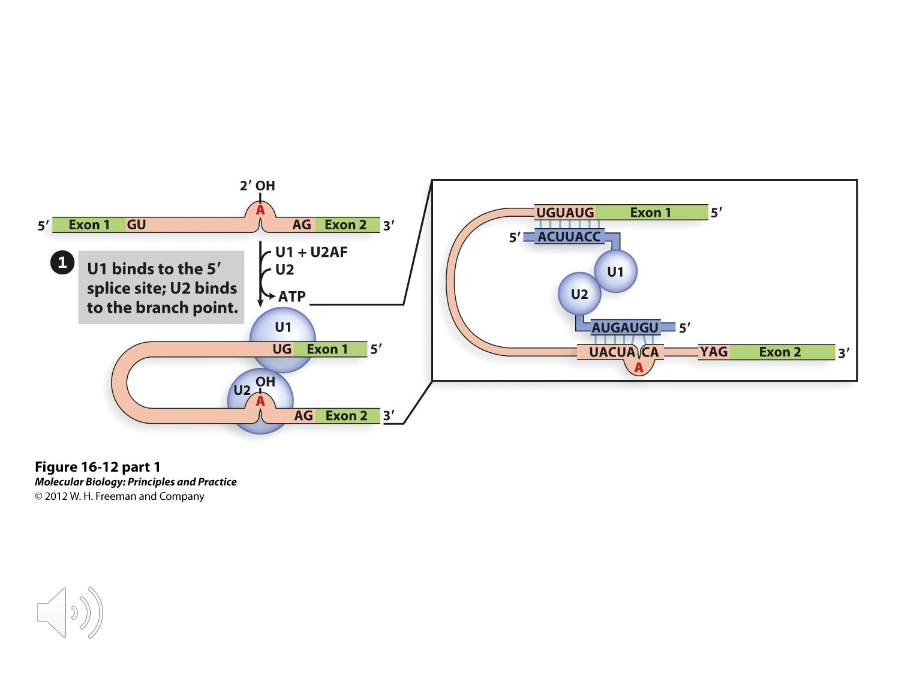
U1/U2
The ( ) protein recognizes the GU at the 5’ splice site while U2AF binds to the AG at the 3’ splice site. This recruits ( ) to the branch point.
U4-U5-U6 Complex
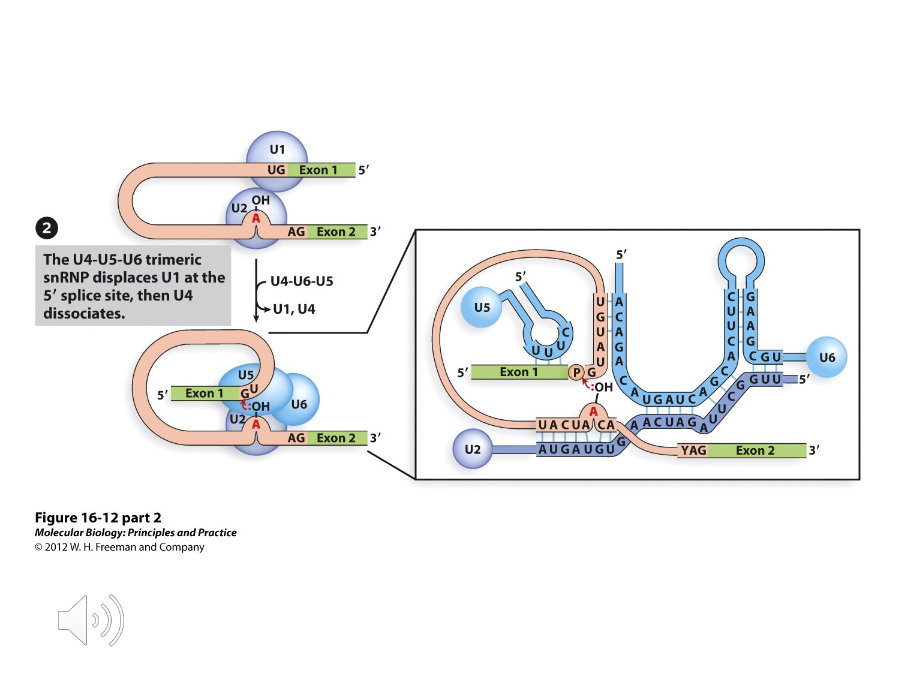
A complex of proteins that binds to and replaces U1 at the 5’ splice site. U6 will bind to U2 and U4 is released.
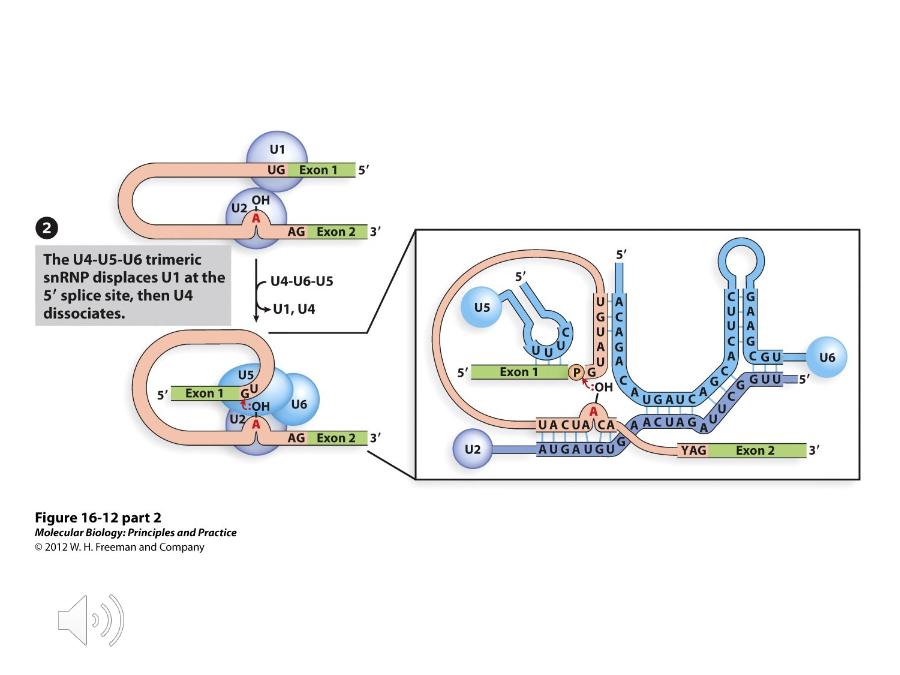
Branch point
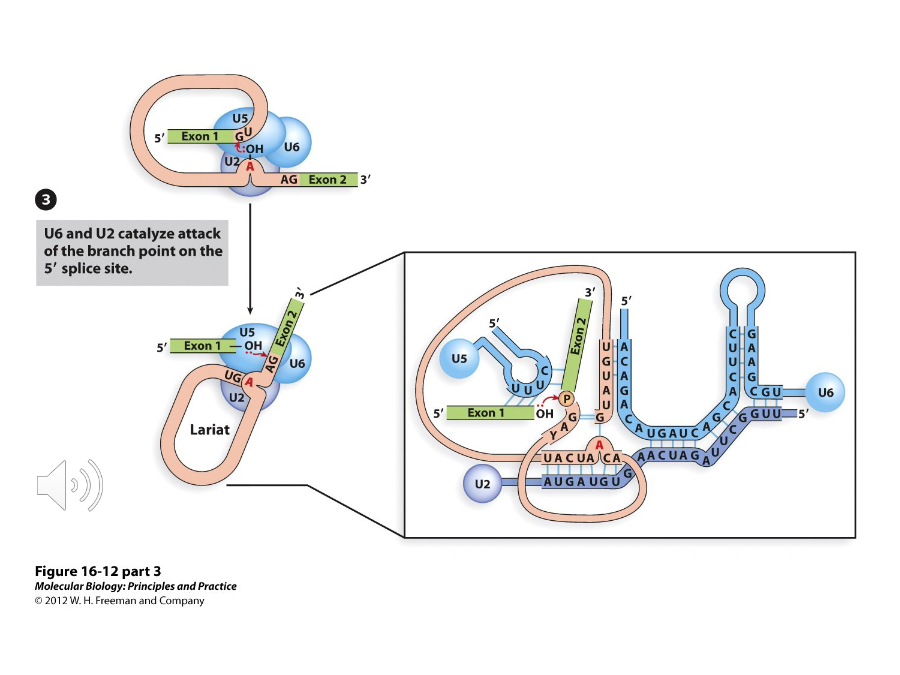
Once the U4-U5-U6 complex attacks, the 2’ OH group of the A at the ( ) attacks the 5’ splice site, which is catalyzed by U6 and U2.
U5
A protein in the U4-U5-U6 complex that base pairs with the 5’ exon and the 3’ splice sites. Because of this, the exons are joined together and the intron is liberated when the 3’ splice site is attacked.
Self-Splicing Introns
Introns capable of removing themselves without any help.There exists two groups of these: Group I and Group II.
Group I Introns
Self-splicing introns that splice by a totally different pathway. Instead of an adenine in the branch point, they use a free guanine nucleotide and its 3’ OH group to attack the 5’ splice site, adding G to the intron and breaking off the exon. This exon then attacks the 3’ splice site.
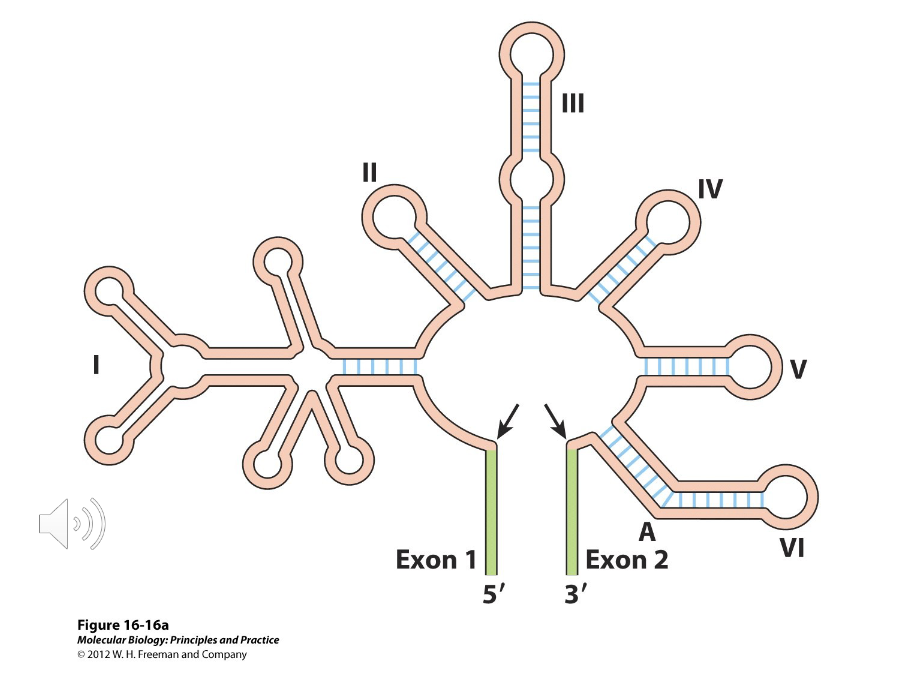
Group II Introns
Self-splicing introns whose chemistry of splicing and produced RNA intermediates are the same as they are in nuclear splicing.
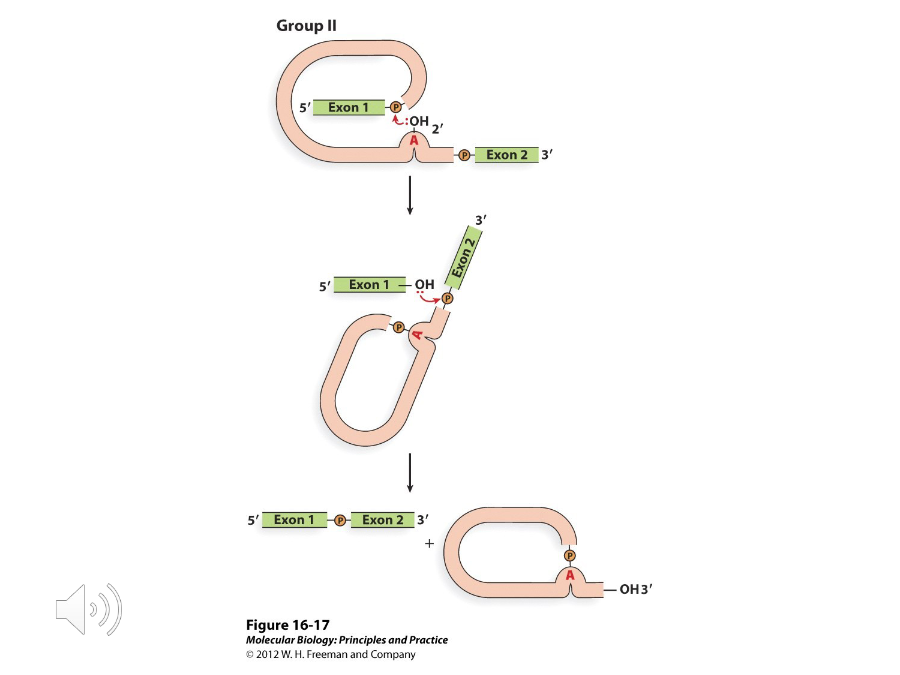
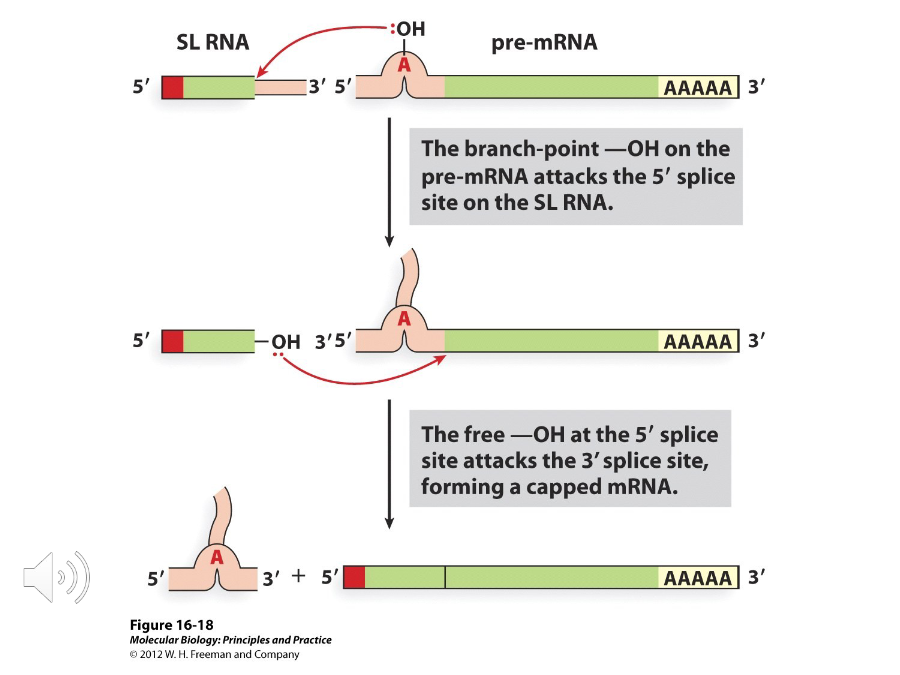
Trans Splicing
When two exons from different transcripts are joined together using the same chemistry as nuclear introns.