Genetics Exam 3
1/86
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
87 Terms
Negative supercoiling
The chromosomal DNA in bacteria is negatively supercoiled.
Negative supercoiling has two major effects:
1. Helps in the compaction of chromosome
2. In localized regions, creates tension that may be released by DNA strand separation
positive supercoiling
Positive supercoiling serves to compact DNA, regulate gene expression and coordinate the transcriptional response to environmental signals.
DNA gyrase
DNA gyrase introduces negative supercoils using energy from ATP, can relax positive supercoils, untangle intertwined DNA .
DNA topoisomerase I
relaxes negative supercoils
Eukaryotic chromosomes
Each chromosome contains a single, linear molecule of DNA, contain multiple origins of replication
Centromeres
regions that play a role in segregation of chromosomes
telomeres
specialized regions at the ends of chromosomes
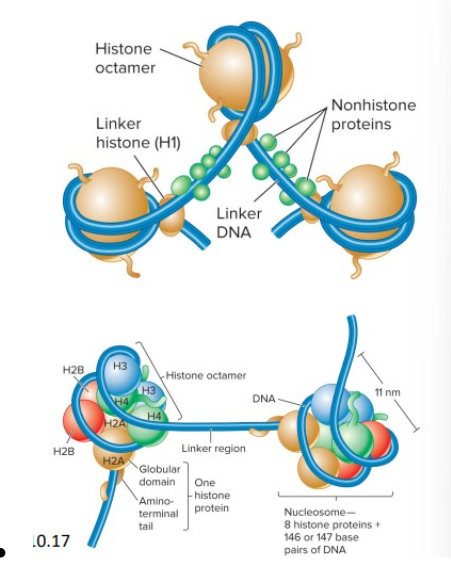
composition and function of a nucleosome
DNA is packaged around histone proteins to form nucleosomes. Within the chromatin and composed of a double-stranded segment of DNA wrapped around an octamer (composed of two copies each of four different histone proteins), associate with each other to form a more compact structure.
Histone proteins
basic, contain many positively-charged amino acids (lysine and arginine), bind to the negatively-charged phosphates along the DNA backbone
Five types of histones
H2A, H2B, H3 and H4 are the core histones
▪ H1 is called the linker histone (binds to DNA in the linker region, less tightly bound to DNA than core histones, helps to organize adjacent nucleosomes)
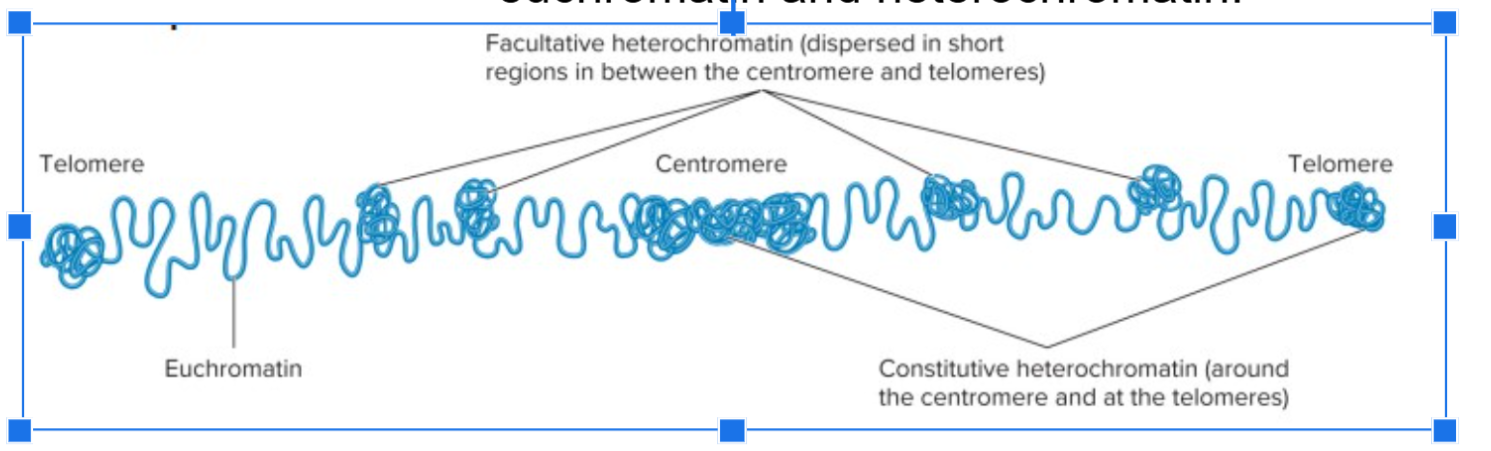
euchromatin
less condensed regions of chromosomes, transcriptionally active, loop domains are less compacted.
heterochromatin
tightly compacted regions of chromosomes, transcriptionally inactive, loop domains are more compacted.
constitutive heterochromatin
regions that are always heterochromatic, permanently inactive for transcription, and usually contain highly repetitive sequences.
facultative heterochromatin
regions that can interconvert between euchromatin and heterochromatin.
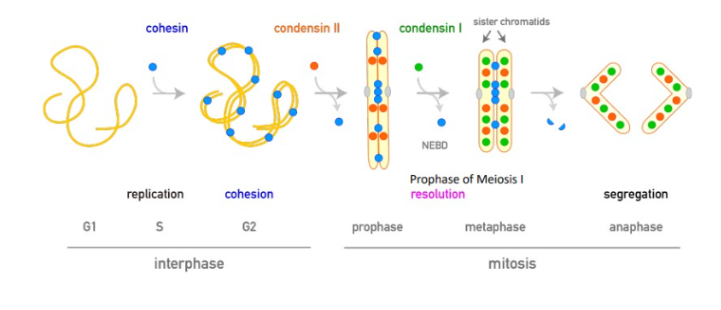
cohesin
a multiprotein complex that also contains SMC proteins SMC1+SMC3, promoting binding between sister chromatids during mitosis and meiosis, only found at the centromere and degraded during anaphase.
condensin
compacts chromosomes during mitosis and meiosis. Both condensin I and II facilitate the reorganization of chromosomes into radial loop arrays.
transposition
involves the integration of small segments of DNA into a new location in the genome.
Small, mobile DNA segments are termed transposable elements (TEs) - Barbara McClintock.
simple transposons
Used widely by transposable elements, or transposons, TE is removed from its original site and transferred to a new target site.
Mechanism is called a cut-and-paste mechanism because the element is cut out of its original site and pasted into a new one.
direct repeats (DRs)
all TE are flanked, which are identical base sequences that are oriented in the same direction and repeated.
insertion element
simplest TE, which is flanked by inverted repeats
inverted repeats
DNA sequences that are identical but run in opposite directions
transposase
catalyzes the transposition event
autonomous elements
contain all the information necessary for transposition or retrotransposition
nonautonomous element
lacks a gene such as one that encodes transposase or reverse transcriptase, which is necessary for transposition.
retrotransposons
move via an RNA intermediate and are transcribed into RNA, found only in eukaryotic species.
use an RNA intermediate in their transposition mechanism.
LTR retrotransposons
are evolutionarily related to known retroviruses, contain long terminal repeats (LTRs)
LTR retrotransposon movement requires two key enzymes:
Reverse transcriptase uses this RNA as a template to synthesize a double-stranded DNA molecule.
Integrate recognizes LTRs at the ends of the DNA, makes cuts at a target site in the host chromosome, and catalyzes the insertion of the TE into the site
Non-LTR retrotransposons
do not resemble retroviruses in having LTR sequences, moved by target-site primed reverse transcription.
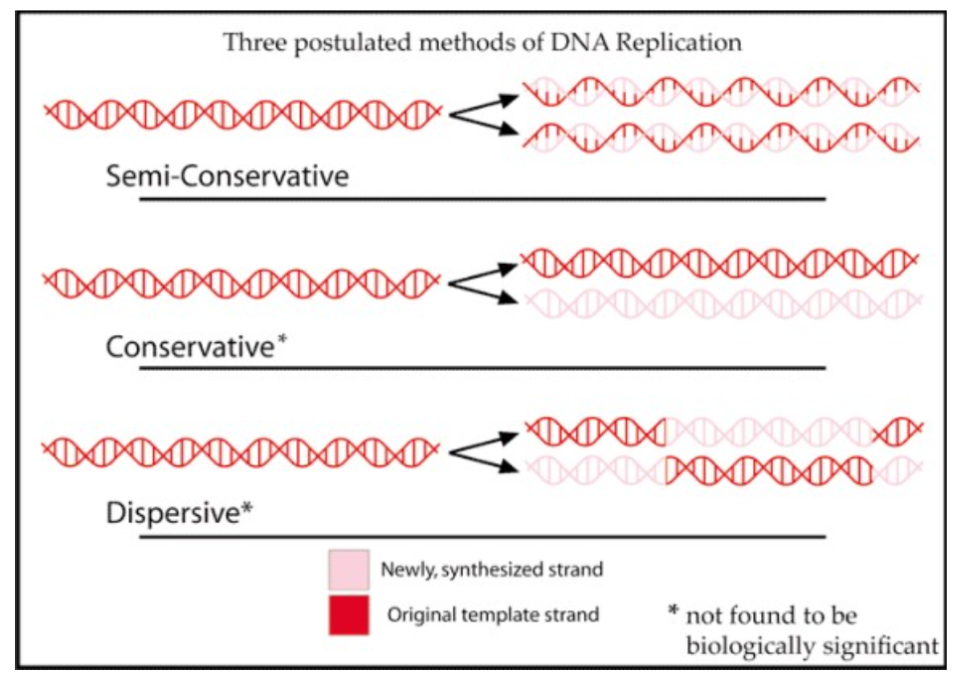
Meselson’s and Stahl’s
by using a density-labeling technique that showed DNA replication was semi-conservative.
DNA helicase
binds to the origin, breaks the hydrogen bonds between the DNA strands, further separates the DNA strands, composed of six subunits, travels along the DNA in the 5’ to 3’ direction, uses energy from ATP
Primase
short RNA primers being synthesized
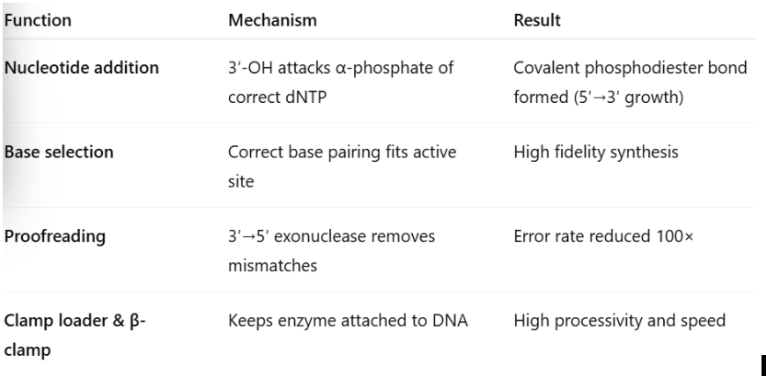
DNA polymerase
are the enzymes that catalyze the attachment of nucleotides to synthesize a new DNA strand.
DNA pol III
normal replication, responsible for most of the DNA replication, processive enzyme due to several different subunits in the DNA pol II holoenzyme, synthesizes a daughter strand of DNA
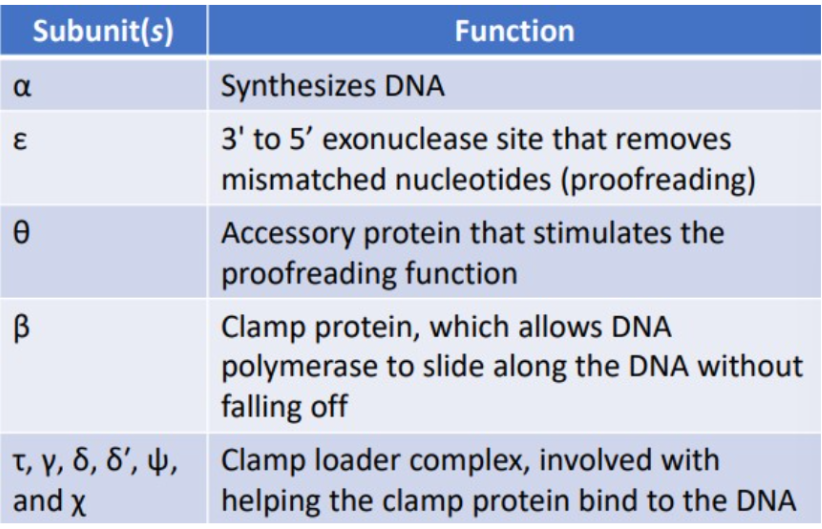
DNA pol I
normal replication, removes the RNA primers and replaces them with DNA, uses a 5’ to 3’ exonuclease activity to digest the RNA and 5’ to 3’ polymerase activity to replace it with DNA
DNA ligase
catalyzes the formation of a covalent (ester) bond to connect the DNA backbones, covalently links the DNA backbones in the Okazaki fragments together
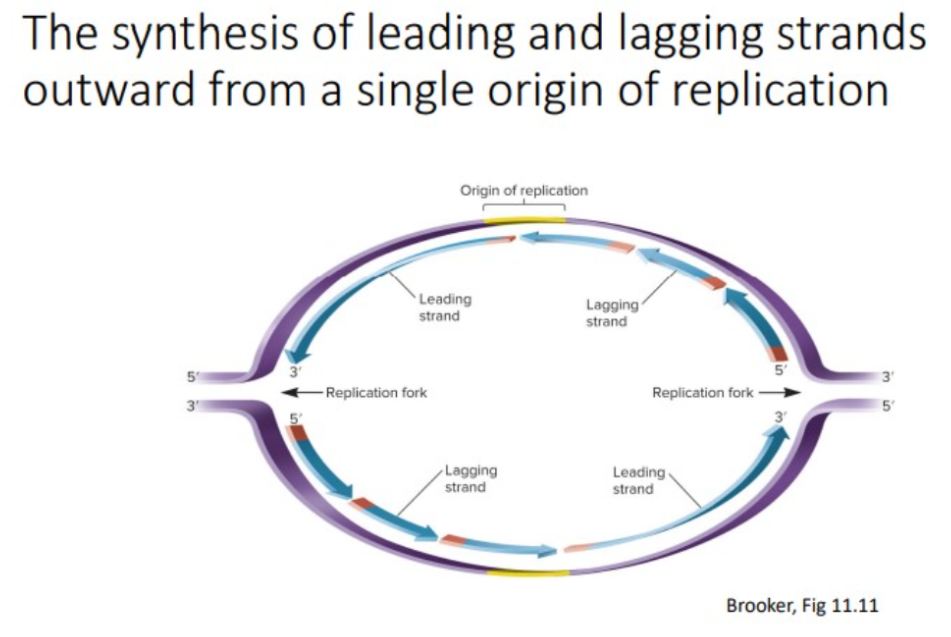
leading strand
one primer is made at the origin, DNA pol III attaches nucleotides in a 5’ to 3’ direction as it slides toward the opening of the replication.
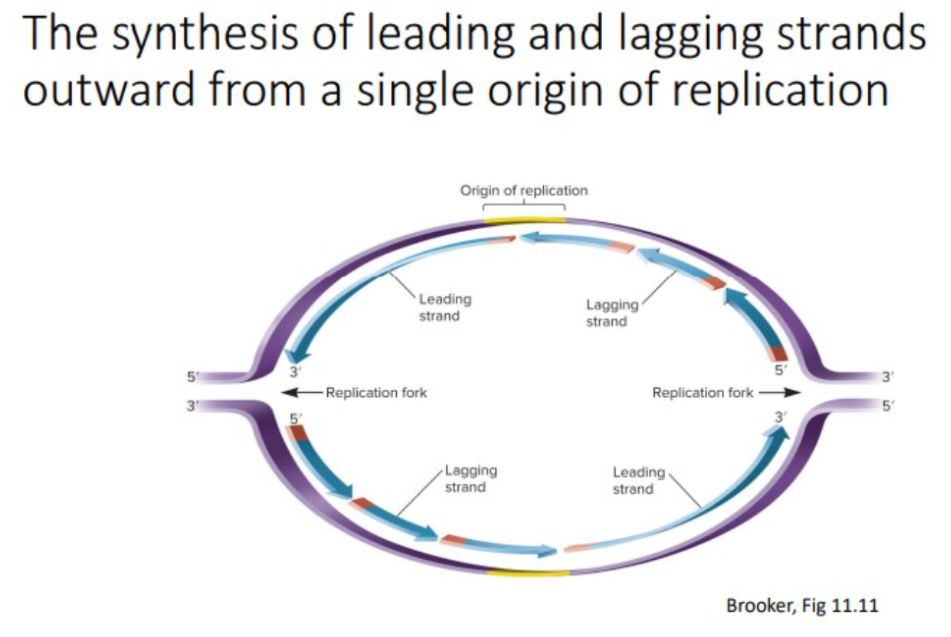
Lagging strand
synthesis is also in the 5’ to 3’ direction, away from the replication fork; many RNA primers are required; DNA pol III uses the RNA primers to synthesize small DNA fragments.
Explain how DNA polymerase III attaches nucleotides to a growing DNA strand while also decreasing the error rate during replication.
Explain why telomerase is needed for replication of the ends of eukaryotic chromosomes.
1. Binding 2. Polymerization 3. Translocations are steps repeated many times to preserve the length of one strand.
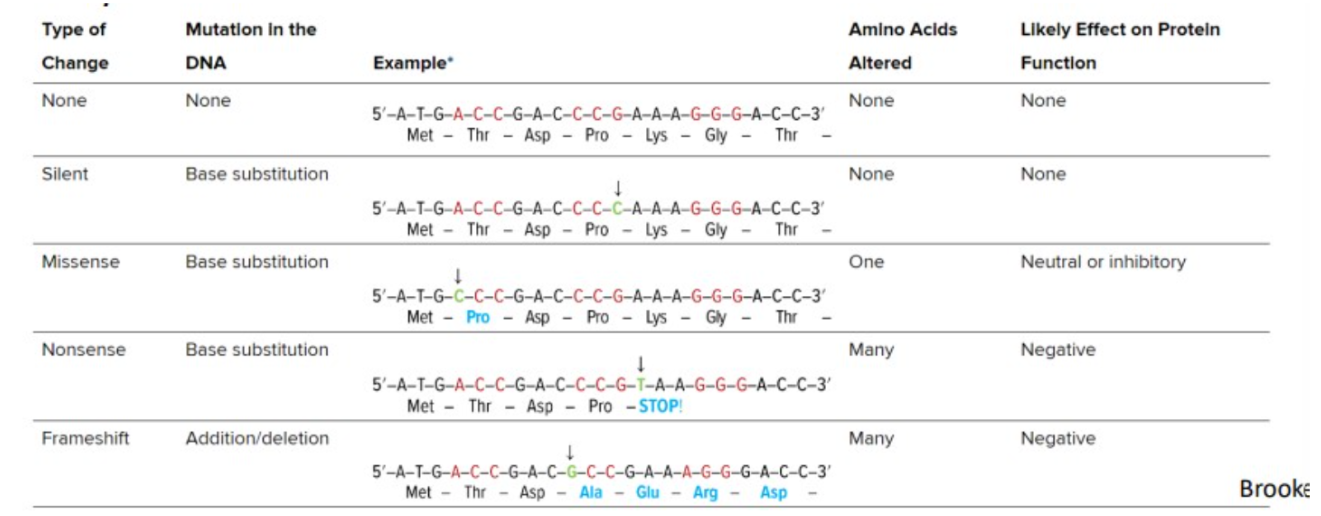
silent mutations
base substitutions that do not alter the amino acid sequence of the polypeptide
missense mutations
base substitutions in which an amino acid change does occur
nonsense mutations
base substitutions that change a normal codon to a stop codon
frameshift mutations
addition or deletion of a number of nucleotides that is not divisible by three
Describe how a mutation within the coding sequence of a gene may alter a polypeptide’s structure and function.
Intragenic
suppressors a second mutation in the same gene restores function
Intergenic
a different gene that counteracts the detrimental effects of a mutation in a first gene
depurination
the removal of a purine (guanine or adenine) from the DNA to form an apurinic site, which can be repaired by DNA repair pathways.
deamination
the removal of an amino group from the cytosine base; C to U/T mutations, can be repaired by DNA repair pathways
Tautomeric shift
a temporary change in base structure; the common, stable form of thymine and guanine is the keto form (enol); the common, stable form of adenine and cytosine is the amino form (imino)
Base excision repair (BER)
involves a category of enzymes known as DNA N-glycosylases (eliminates abnormal bases)
Nucleotide excision repair (NER)
type of system can repair many types of DNA damage, including thymine dimers and chemically modified bases, missing bases, some types of crosslinks
mismatch repair system
recognize and correct a base pair mismatch; DNA polymerases have a 3’ to 5’ proofreading ability that can detect base mismatches and fix them
Specific to the newly made strand (occurs right after replication)
In E. coli, three proteins, MutL, MutH and MutS detect the mismatch and direct its removal from the newly made strand
MutH distinguishes between the parental strand (methylated) and the daughter strand (not methylated)
Promoter
DNA site for RNA polymerase to bind; signals start of transcription
Terminator
DNA site signaling end of transcription
activator
bind to DNA and increase transcription (positive control)
repressor
bind to DNA and inhibit transcription (negative control)
mRNA
the "messenger" carrying genetic code from DNA to the ribosome
rRNA
the structural and catalytic component of the ribosome
tRNA
the "transfer" molecule that brings specific amino acids to the ribosome
inducible
Those that increase transcription of inducible genes are termed inducers; May bind to activators and cause them to bind to DNA. May bind to repressors and prevent them from binding to DNA. (cAMP-CAP, lac operon)
Those that inhibit transcription of repressible genes include:
Corepressors which bind to repressors and cause them to bind to DNA.
Inhibitors which bind to activators and prevent them from binding to DNA. (trp operon)
lactose is absent
a repressor protein binds to the operator, blocking transcription
lactose is present
it acts as an inducer by binding to the repressor, causing it to release and allowing transcription to occur
catabolite activator protein (CAP)
Glucose levels are regulated by the catabolite activator protein (CAP); when glucose is low, CAP binds to the CAP site and stimulates transcription, ensuring the bacteria uses lactose only when glucose is scarce.
trp repressor
When tryptophan levels are low, tryptophan does not bind to the trp repressor, preventing the repressor protein from binding to the operator site.
attenuation
Another mechanism of regulation is attenuation. When attenuation occurs, the RNA is transcribed only to the attenuator sequence, and then transcription is terminated
GTFs
bind to DNA and help position RNA polymerase to begin the process of creating an RNA copy of a gene (transcription).
TFIID
Composed of TATA-binding protein (TBP) and other TBP-associated factors (TAFs); Recognizes the TATA box
TFIIA
Binds to TFIID; Promotes TFIID binding to the TATA box
TFIIB
Binds to TFIID; Enables RNA pol II to bind to core promoter; Also promotes TFIIF binding
TFIIF
Binds to RNA pol II; Plays role in RNA pol II’s ability to bind TFIIB and the core promoter; Plays role in ability of TFIIE and TFIIH to bind to RNA pol II
TFIIE
Role in the formation or maintenance (or both) of the open complex; May exert effects by facilitating binding of TFIIH to RNA pol II and regulating the activity of TFIIH
TFIIH
Multisubunit protein; Multiple roles
Identify the functions of the components of the pre-initiation complex for eukaryotic transcription.
GTFs
RNA Pol II - enzyme, catalyzes linkage of nucleotides in the 5’ to 3’ direction, using DNA as the template
Mediator - Multisubunit complex; Mediates effects of regulatory transcription factors on RNA pol II; Subunits vary depending on cell type and environmental conditions; Plays key role in RNA pol II switch from initiation to elongation
5’ cap
Recognized by cap-binding proteins which play roles in:
Movement of RNAs out of the nucleus
Early stages of translation
Splicing of introns.
PolyA tail
NOT coded in the gene sequence; Added enzymatically after the gene is completely transcribed; A long PolyA tail is important for mRNA stability, Export from the nucleus, Synthesis of polypeptides.
Introns
(intervening sequences) are removed
Exons
(coding sequences) are connected together
Presence of splicing factors
exons that are recognized and joined together, resulting in one or more specific splice variants, that bind to the spliceosome (a complex of RNA and proteins).
absence of splicing factors
outcome is unpredictable; a different set of exons may be included or excluded, leading to a different splice variant and the spliceosome may bind to incorrect splice sites or not assemble at all.
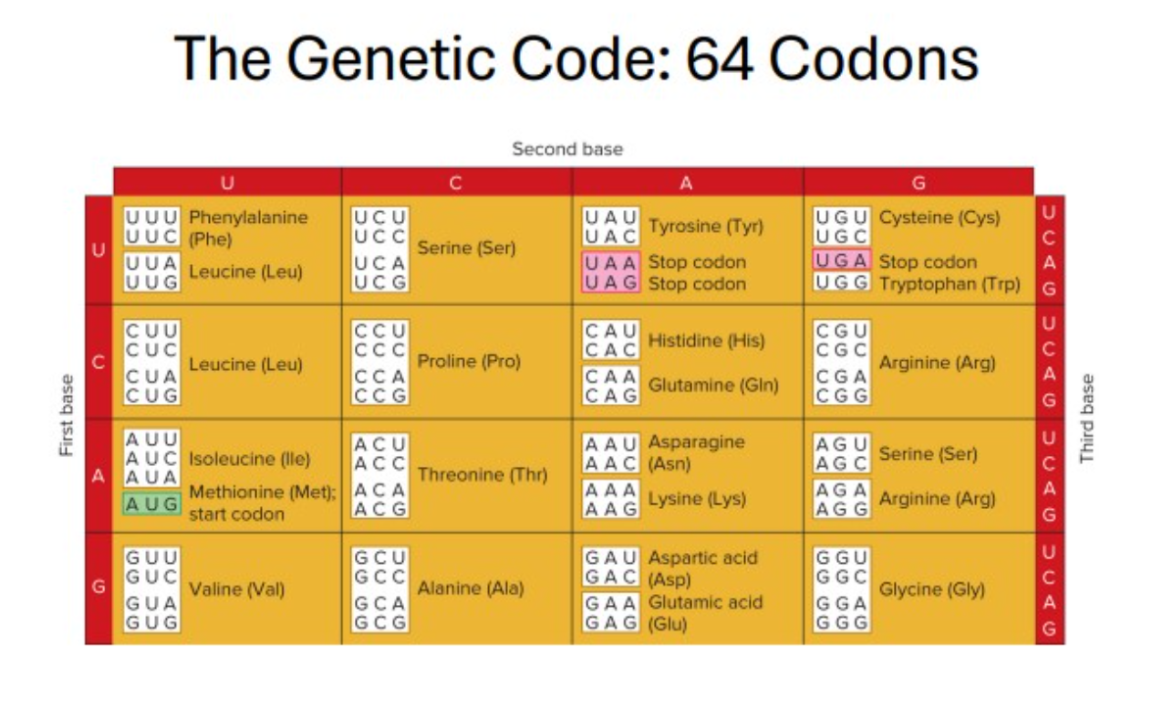
start codon
AUG (which specifies methionine) = start codon
Defines the reading frame for all following codons
Example: 5’ - AUG CCC GGA GGC ACC GUC CAA U - 3’
Met - Pro - Gly - Gly - The - Val - Gln
stop codons
UAA, UAG and UGA = termination, or stop codons
Example: GGU, GGC, GGA, and GGG all code for glycine (are synonymous)
Translate a mature mRNA sequence into a polypeptide chain (using a provided codon table)
four levels of structure in proteins
Primary - amino acid sequence linked by peptide bonds
Secondary - the primary structure of a protein folds to form regular, repeating shapes (a helix, b sheet); stabilized by the formation of hydrogen bonds between atoms located in the polypeptide backbone.
Tertiary - the short regions of secondary structure in a protein fold into a three-dimensional polypeptide; Structure determined by hydrophobic and ionic interactions as well as hydrogen bonds and van der Waals interactions.
Quaternary - proteins made up of two or more polypeptides associate with one another to make a functional protein
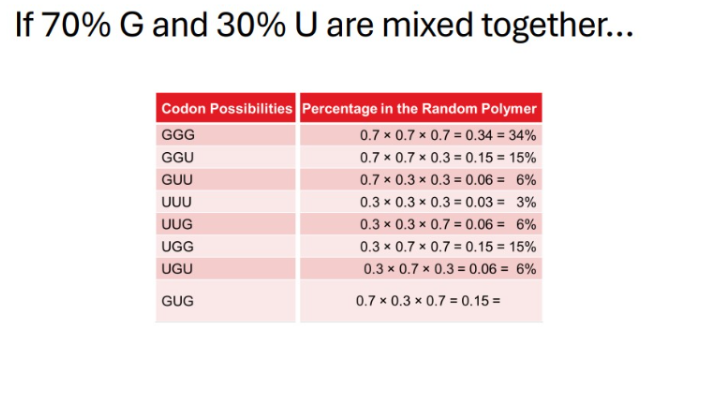
Calculate the expected percentage of a radiolabeled amino acid from a cell-free translation experiment
Describe the functions of the 16S, 30S, and 50S subunits of the bacterial ribosome, identifying any key sites within them.
16S - can detect when an incorrect tRNA is bound at the A site and will prevent elongation until the mispaired tRNA is released from the A site, decoding function. codon-anticodon recognition, Shine-Dalgarno binding
30S - decoding, initiation, mRNA binding (A site, P site)
50S - peptide bond formation, elongation, peptide exit (peptidyl transferase, E site, exit tunnel)
Explain what’s happening in the ribosome during the elongation stage of translation.
1. A charged tRNA brings a new amino acid to the ribosome so that it can be attached to the end of the growing polypeptide and binds to the A site
2. Next step of elongation is peptidyl transfer—the polypeptide is removed from the tRNA in the P site and transferred to the amino acid at the A site
3. The ribosome then moves, or translocates, to the next codon in the mRNA, moving the tRNAs at the P and A sites to the E and P sites
4. Uncharged tRNA is released from the E site.
5. Repeat steps 1-4 until a stop codon appears