2.1.3 Nucleotides & Nucleic acids
1/16
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
17 Terms
Nucleotides
Nucleotides are the monomers from which nucleic acids, like DNA & RNA are formed
Nitrogenous bases can be categorised according to their ring structure. The 2 categories are purines and pyrimidines
Purines - 2 carbon ring structure (Adenine & Guanine)
Pyrimidines - 1 carbon ring structure(cytosine, thymine, uracil)
DNA
T-A
G-C
RNA
T change to U
DNA & RNA
Both DNA & RNA undergo condensation reactions, forming phosphodiester bonds between the nucleotide monomers, to create the polymer. The polymer of these nucleotides is called a polynucleotide. A phosphodiester bond is a strong covalent bond that forms between the pentose sugar & phosphate of different nucleotides.
ATP
Adenosine Triphosphate
contains 3 phosphate ions
essential for metabolism
is an immediate source of energy for biological processes
ATP is compos of adenine ribose (pentose sugar) and 3 inorganic phosphate groups
Made during respiration via a condensation reaction and the enzyme ATP synthase
ADP + Pi → ATP + H2O
ATP + H2O → ADP + Pi (ATP is hydrolysed using the enzyme ATP hydrolase
DNA (Deoxyribonucleic Acid)
codes for the sequence of amino acids in the primary structure of a protein, which determines the final 3D structure & function of the protein.
Double helix of 2 anti parallel strands join together by H bonds between the bases on the 2 different sides.
How DNA structure relates to its function
→stable structure due to the sugar-phosphate backbone (covalent bonds) and the double helix
→Double-stranded so replication can occur using both strands as a template
→Weak hydrogen bonds between the bases for easy separation of the 2 strands in a double helix during replication
→A large molecule that carries a lot of information
→Complementary bases pairing allows identical copies to be made
DNA precipitation
-Homogenise → with detergent. Break open the cells and membranes to release contents.
-Filter to remove debris
-Add salt to break H bonds between DNA + water molecules
-Add protease to digest proteins
-Add ice-cold ethanol to precipitate out the DNA. The DNA appears as white strands
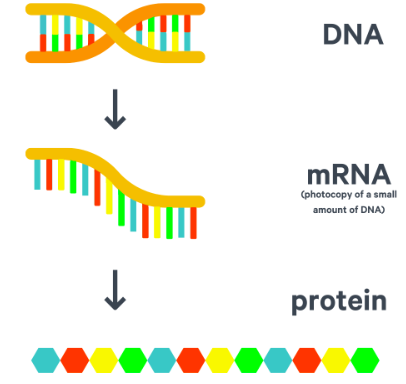
mRNA
-A copy of one gene from DNA
-Created in the nucleus and it then leaves via the nuclear pore to carry the copy of the genetic code of one gene to a ribosome in the cytoplasm
-mRNA is shorter than DNA
-mRNA is short-lived
-mRNA is single-stranded & every 3 bases in the sequence code for one specific amino acid these 3 bases are therefore codons
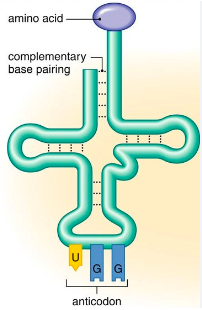
tRNA
-Found in the cytoplasm
-It is single-stranded but folded to create a cloverleaf shape held in place by H bonds
-Brings a specific amino acid to the ribosome
-This is determined by 3 bases found on the (tRNA) anticodon, which are complementary to the 3 bases on the mRNA (codon)
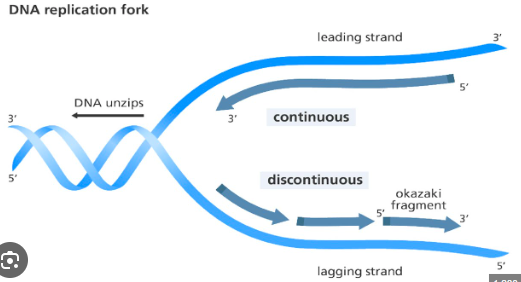
How DNA is replicated
First the enzyme gyrase will unwind the double helix structure once the helix is unwound, the enzyme DNA helicase will unzip the 2 strands by breaking the hydrogen bonds. Free nucleotides will come along and pair with the unpaired bases. With the help of DNA polymerase, Phosphodiester bonds form between nucleotides. In a replication fork continuous replication→ DNA polymerase helps join the free nucleotides from 5’ to 3’ prime end creating a leading strand. Discontinuous replication, free nucleotides are added to unpaired bases. DNA polymerase can only have from 5’ to 3’ prime end which creates a lagging strand. Nucleotides are bonded by phosphodiester bonds, DNA Ligase binds the Okazaki fragments together.
Semi Conservative DNA Replication
-one strand is conserved and one new strand is created
-copying errors in DNA can occur randomly and spontaneously, resulting in a change to the DNA base sequence, known as a mutation
-DNA replication occurs in S-phase in interphase of the cell cycle
-When describing the DNA double helix, the top and bottom of each strand are described as either the 3’ prime end or the 5’ prime end. The enzyme that catalyses DNA replication is complementary in shape to the 3’ end & can therefore only attach to the DNA at this location.
DNA replication fork
The Genetic Code
-Degenerate - Amino Acids are coded for by more than 1 triplet of bases
-Universal - The same triplet of bases codes for the same Amino Acid in all organisms
-Non-overlapping - each base in a gene is only part of 1 triplet of bases that codes for 1 amino acid. Each codon, or triplet of bases is read as a discrete unit.
Protein synthesis
1) Transcription → where the DNA sequence for one gene is copied into mRNA
2) Translocation → where the mRNA joins with a ribosome and a corresponding tRNA molecule brings the specific amino acid the codon for.
Transcription
1) DNA section containing the gene is unwound using enzyme gyrase and H bonds broken → unzipped - Helicase
2) Free RNA nucleotides hydrogen bond to complementary bases on the template strand (uracil replaces thymine)
3) RNA polymerase catalyses the formation of phosphodiester bonds between RNA nucleotides (5’→3’)
4) Once RNA polymerase reaches the stop codon, the pre-mRNA DNA returns to a double helix shape
5) Before translation, splicing occurs → introns - non-coding regions are removed
→extrons - coding regions stick together
Translocation
6) In the cytoplasm mRNA joins with a ribosome at the start codon
7) A tRNA molecule with a complementary anticodon joins to the codon (with H-bond) to the mRNA, each codon codes for a specific amino acid on the tRNA to start it = methionine (AUG=codon , UAC =anticodon)
8) Ribosome moves along the mRNA + another tRNA molecule joins each carrying different amino acids
9) Enzyme + ATP used to join the amino acids together forming a peptide bond
10) This repeats along the mRNA, tRNA molecules leave amino acid behind
11) A stop codon, polypeptide is released + folds into a protein