BIO 130 term 1
5.0(2)
Card Sorting
1/147
Earn XP
Description and Tags
Study Analytics
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
148 Terms
1
New cards
Cell theory(week 1: section 1)
* cell is the basic organizational unit of life
* all organisms are comprised of 1 or more cells
* cells arise from pre-existing cells
* all organisms are comprised of 1 or more cells
* cells arise from pre-existing cells
2
New cards
Prokaryotic cells(week 1: section 1)
* no nuclei
* single-celled
* no membrane-bound organelles
* DNA bound by nucleoid(not membrane)
* smaller than eukaryotes
* less DNA than eukaryotes
* Bacteria and Archaea
* single-celled
* no membrane-bound organelles
* DNA bound by nucleoid(not membrane)
* smaller than eukaryotes
* less DNA than eukaryotes
* Bacteria and Archaea
3
New cards
Eukaryotic cells(week 1: section 1)
* nuclei
* single-celled/multicellular
* several membrane bound organelles
* plants, fungi, animals, humans
* single-celled/multicellular
* several membrane bound organelles
* plants, fungi, animals, humans
4
New cards
Differences between animal and plant eukaryotic cells(week 1: section 1)
* plants are larger and more complex
* plants: cell wall, chloroplasts, large vacuole
* plants: cell wall, chloroplasts, large vacuole
5
New cards
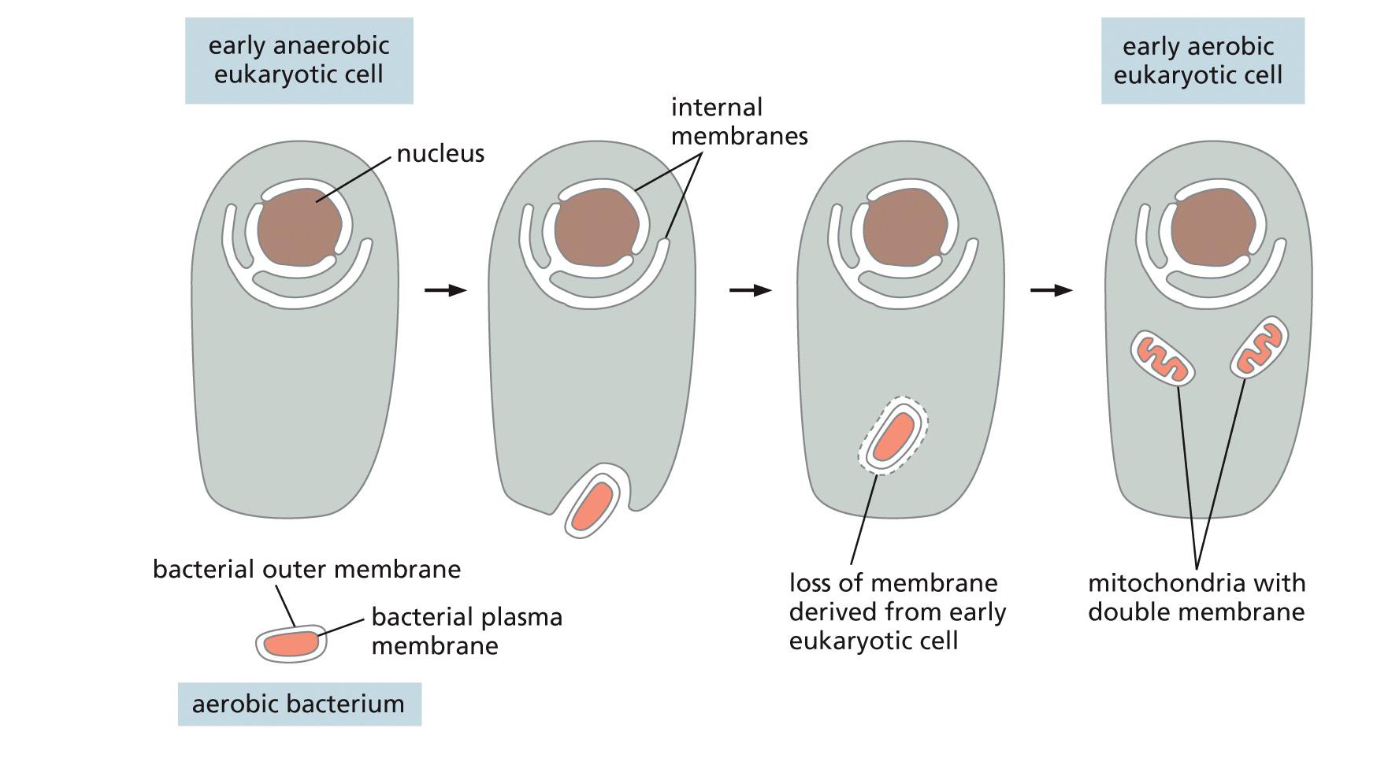
Origins of mitochondria(week 1: section 1)
* aerobic bacterium engulfed by anaerobic eukaryotic cell
* aerobic bacterium loss plasma membrane and split into mitochondria w/ double membrane
* becomes early aerobic eukaryotic cell
* aerobic bacterium loss plasma membrane and split into mitochondria w/ double membrane
* becomes early aerobic eukaryotic cell
6
New cards
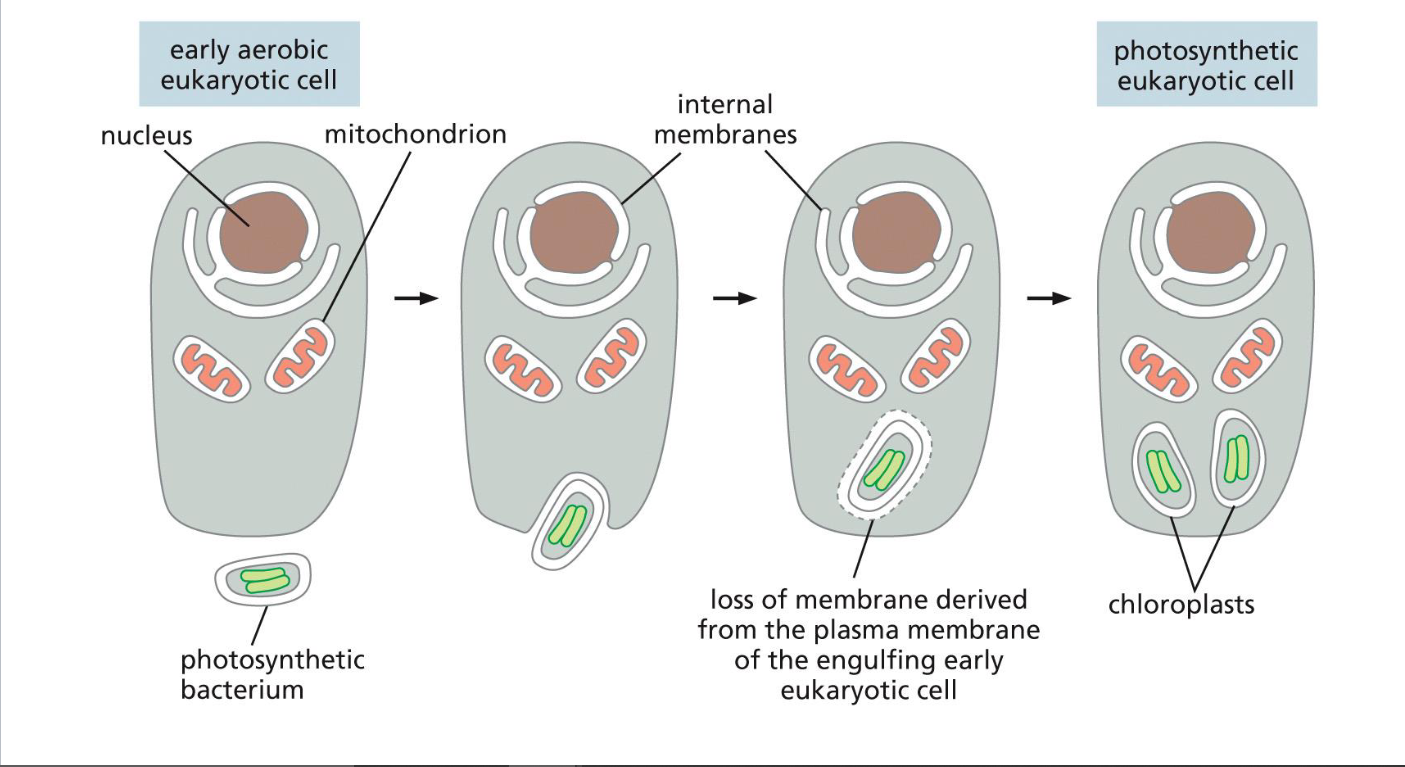
origins of chloroplasts(week 1: section 1)
* early aerobic eukaryotic cell engulfs photosynthetic bacterium
* photosynthetic bacterium loses membrane and splits into chloroplasts
* becomes photosynthetic eukaryotic cell
* photosynthetic bacterium loses membrane and splits into chloroplasts
* becomes photosynthetic eukaryotic cell
7
New cards
Endosymbiont hypothesis(week 1: section 1)
* organelles in eukaryotic cells were once prokaryotic microbes that entered eukaryotic cells living together
* shown w/ origins of mitochondria + chloroplasts
* shown w/ origins of mitochondria + chloroplasts
8
New cards
Evidence for endosymbiont hypothesis(week 1: section 1)
1. remnants of mitochondria + chloroplasts’s genomes + genetic systems resemble that of modern day prokaryotes
2. their own protein + DNA synthesis components resemble prokaryotes too
9
New cards
General attributes of model organisms(week 1: section 2)
* rapid development w/ short life cycles
* small adult(reproductive) size
* readily available(collections or widespread)
* tractability (ease of manipulation or modification)
* understandable genetics
* small adult(reproductive) size
* readily available(collections or widespread)
* tractability (ease of manipulation or modification)
* understandable genetics
10
New cards
Examples of model organisms(week 1: section 2)
* E. coli
* Brewer’s yeast
* Arabidopsis thaliana (wall cress)
* Drosophila melanogaster (fruit fly)
* Caenorhabditis elegans (nematode worm)
* Zebrafish
* mice
* Brewer’s yeast
* Arabidopsis thaliana (wall cress)
* Drosophila melanogaster (fruit fly)
* Caenorhabditis elegans (nematode worm)
* Zebrafish
* mice
11
New cards
E. coli as a model organism(week 1: section 2)
* prokaryote
* bacteria
* helps show fundamental mechanisms of life (ex: how cells replicate)
* bacteria
* helps show fundamental mechanisms of life (ex: how cells replicate)
12
New cards
Brewer's yeast as a model organism(week 1: section 2)
* eukaryote
* single celled __fungus__ similar to plant cells (has cell wall, immobile, no chloroplasts)
* simple eukaryote to help study more complex ones
* single celled __fungus__ similar to plant cells (has cell wall, immobile, no chloroplasts)
* simple eukaryote to help study more complex ones
13
New cards
Arabidopsis thaliana(wall cress) as a model organism(week 1: section 2)
* eukaryote
* weed plant
* helps give insight into development + physiology of crop plants, as well as other plant species
* weed plant
* helps give insight into development + physiology of crop plants, as well as other plant species
14
New cards
Drosophila melanogaster(fruit fly) as a model organism(week 1: section 2)
* eukaryote
* fly (insect)
* helps to understand how all animals develop
* fly (insect)
* helps to understand how all animals develop
15
New cards
Caenorhabditis elegans(nematode worm) as a model organism(week 1: section 2)
* eukaryote
* relative of eel worms
* hermaphrodite
* complete genome (959 body cells)
* share genes w/ humans, so helps to see how humans develop
* relative of eel worms
* hermaphrodite
* complete genome (959 body cells)
* share genes w/ humans, so helps to see how humans develop
16
New cards
zebra fish as a model organism(week 1: section 2)
* eukaryote
* transparent first 2 weeks of life
* helps to see how cells behave during development of a living animal
* transparent first 2 weeks of life
* helps to see how cells behave during development of a living animal
17
New cards
mice as a model organism(week 1: section 2)
* eukaryote
* used to study mammalian genetics, development, immunology + cell bio.
* used to study mammalian genetics, development, immunology + cell bio.
18
New cards
Model organisms + humans(week 1: section 2)
* study of model organisms helps us to understand humans bc:
* humans can also be studied using:
1. clinical studies
2. cell cultures
3. organoids
* humans can also be studied using:
1. clinical studies
2. cell cultures
3. organoids
19
New cards
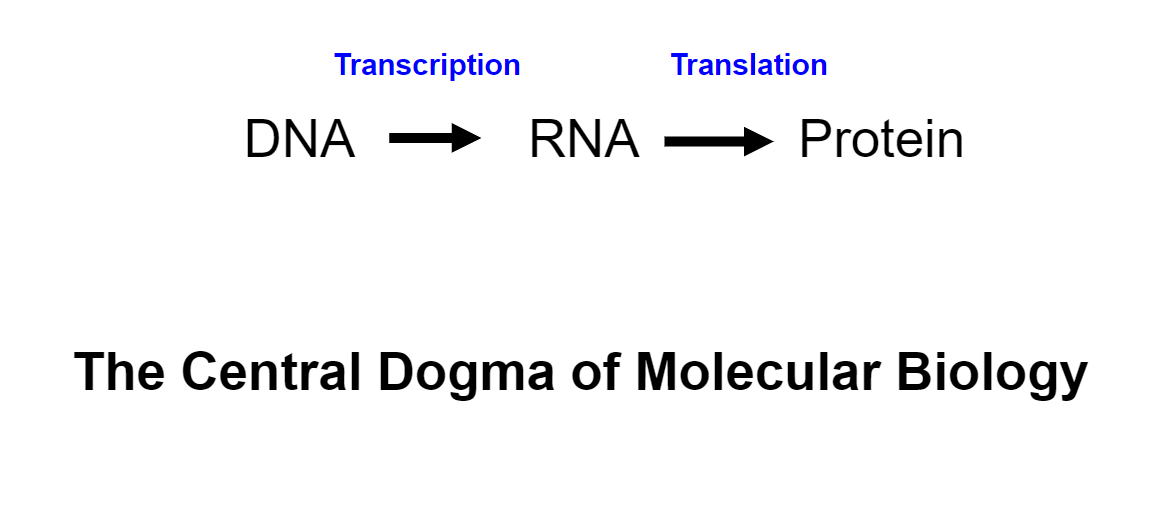
information flow in the cell(week 1: section 3)
\
20
New cards
Refined central dogma(week 1: section 3)
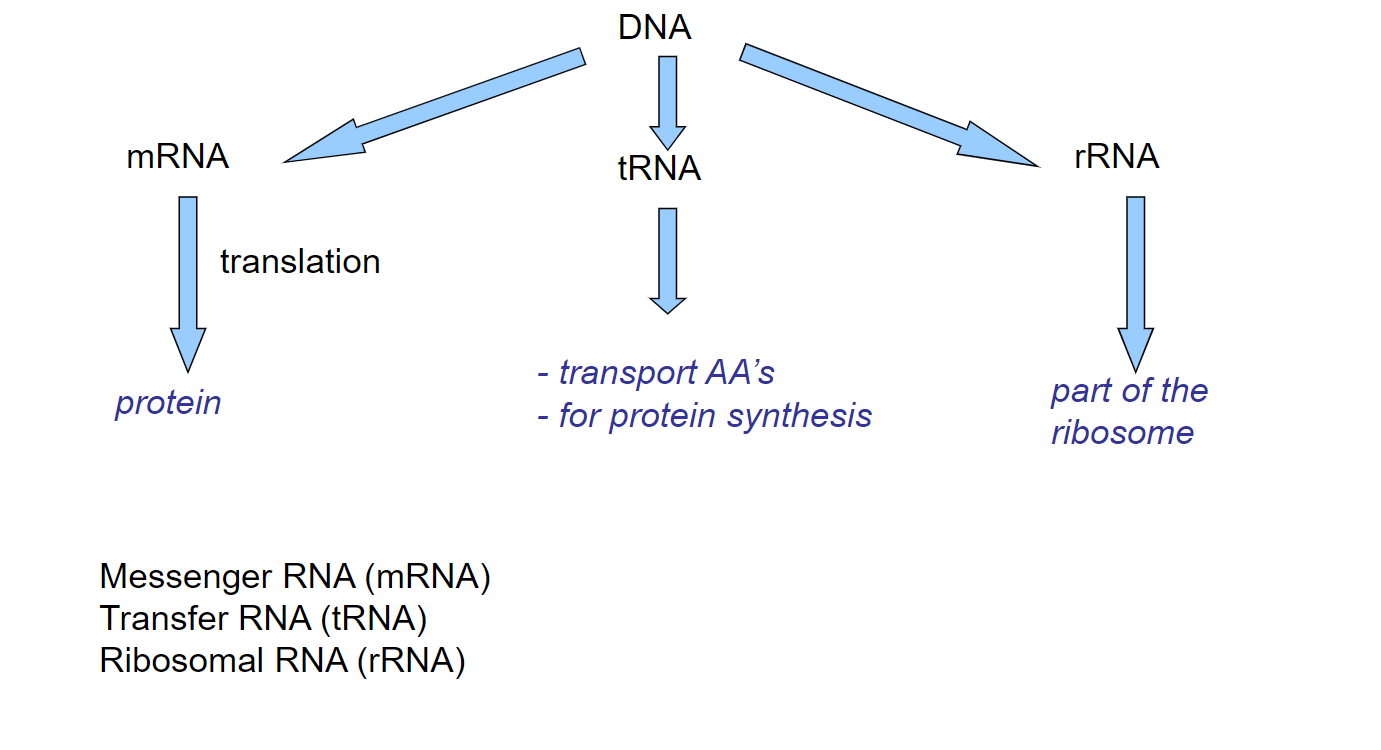
21
New cards
Elaborated central dogma(week 1: section 3)
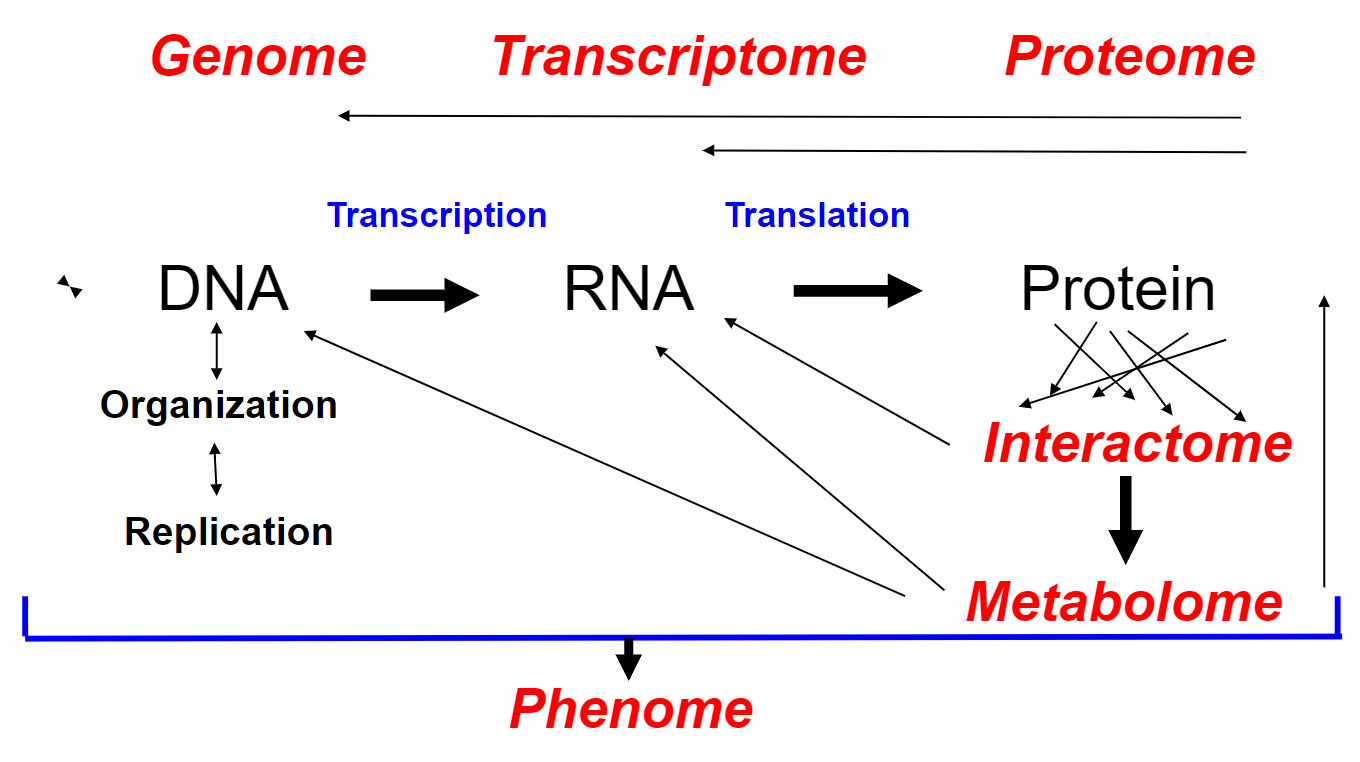
22
New cards
Information flow in prokaryote + eukaryote cells(week 1: section 3)
* DNA, RNA + proteins __synthesized__ as ==linear chains of info w/ a definite polarity==
* info in RNA sequence is translated into amino acid sequence via ==genetic code==(universal among all species)
* info in RNA sequence is translated into amino acid sequence via ==genetic code==(universal among all species)
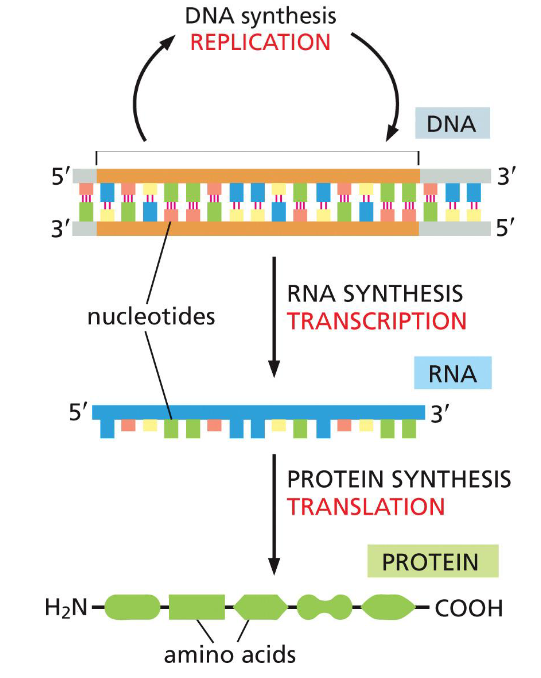
23
New cards
Nucleic acids(week 1: section 4)
* genetic material in a cell(organism’s blueprints)
* DNA = deoxyribonucleic acid
* RNA = ribonucleic acid
* DNA = deoxyribonucleic acid
* RNA = ribonucleic acid
24
New cards
Parts of a nucleotide(week 1: section 4)
1. pentose sugar(foundation for base)
2. nitrogenous base(A, T, C, G, U)
3. phosphate group(backbone, 1-3 P’s)
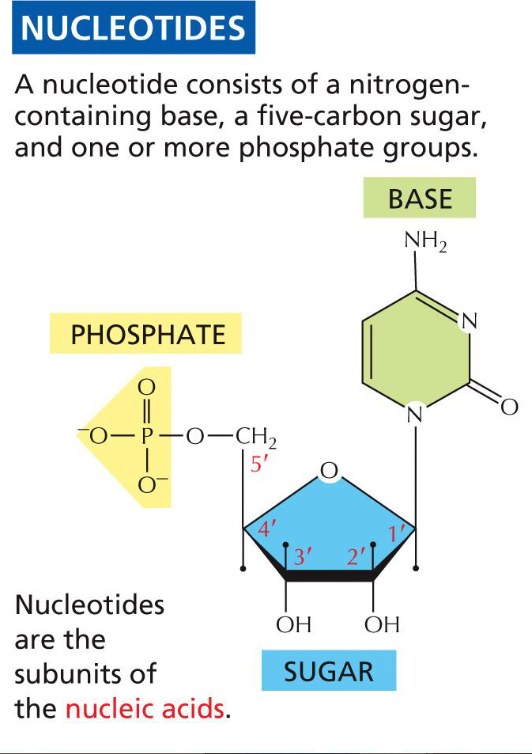
25
New cards
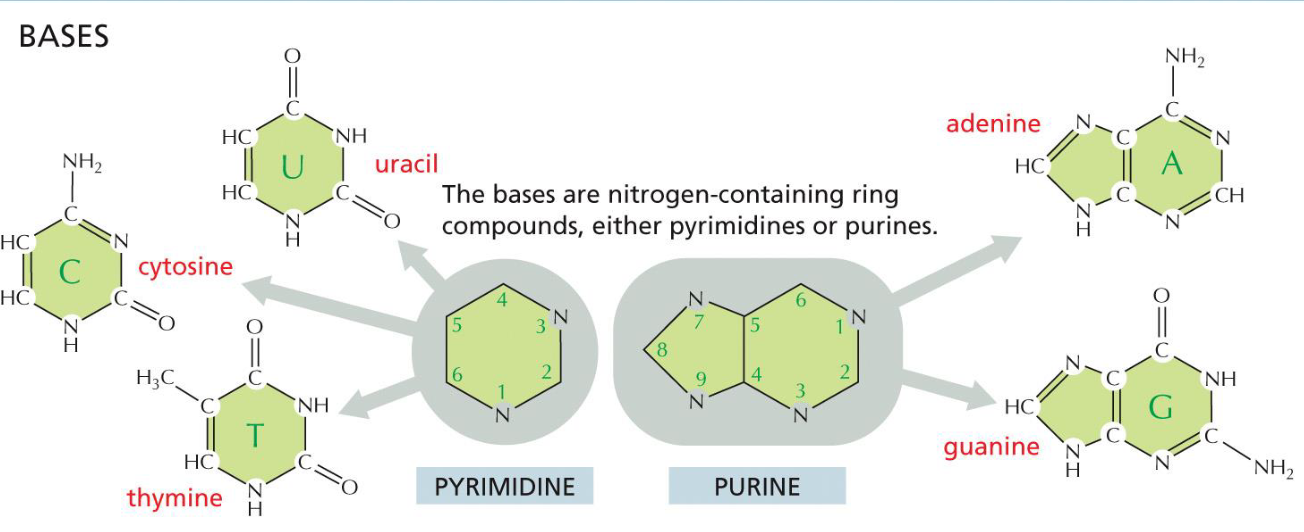
Bases(week 1: section 4)
* nitrogen containing ring compounds
* single ring = pyrimidine (U, T, C)
* double ring = purine (A, G)
* single ring = pyrimidine (U, T, C)
* double ring = purine (A, G)
26
New cards
Differences between RNA and DNA(week 1: section 4)
RNA:
* ribose sugar
* G, C, A, U
DNA:
* deoxyribose sugar(missing oxygen)
* G, C, A, T
* ribose sugar
* G, C, A, U
DNA:
* deoxyribose sugar(missing oxygen)
* G, C, A, T
27
New cards
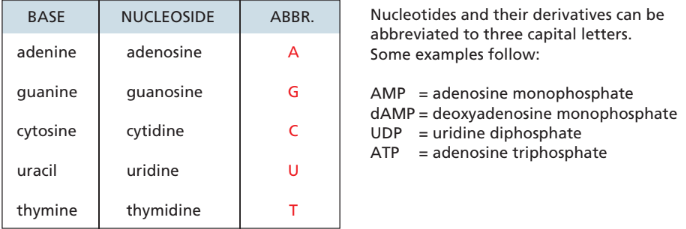
Nucleic acid nomenclature(week 1: section 4)
* base + sugar = nucleo__s__ide
* base + sugar + phosphate = nucleo__t__ide
* 1 phosphate = mono, 2 = di, 3 = tri
* base + sugar + phosphate = nucleo__t__ide
* 1 phosphate = mono, 2 = di, 3 = tri
28
New cards
Bases and their nucleoside naming(week 1: section 4)
29
New cards
Nucleic acid chains(week 1: section 4)
* DNA synthesized from deoxyribonucleoside triphosphates(dNTPs)
* RNA synthesized from ribonucleoside triphosphates(NTPs)
* nucleotides are linked by ==phosphodiester bonds==
* RNA synthesized from ribonucleoside triphosphates(NTPs)
* nucleotides are linked by ==phosphodiester bonds==
30
New cards
Molecular interactions(week 1: section 5)
* interactions btwn individual molecules usually mediated by noncovalent attractions
* individually very weak, but can add up to make strong binding btwn molecules
* individually very weak, but can add up to make strong binding btwn molecules
31
New cards
Types of molecular interactions(week 1: section 5)
1. electrostatic attractions
2. hydrogen bonds
3. van der waals attractions
4. hydrophobic force
32
New cards
electrostatic attractions(week 1: section 5)
* __noncovalent__ force of attraction between 2 oppositely charged molecules
* similar idea to attractions btwn ions or polar molecules
* in bio. can be seen with regions of positive/negative charges on large molecules
* similar idea to attractions btwn ions or polar molecules
* in bio. can be seen with regions of positive/negative charges on large molecules
33
New cards
Hydrogen bond(week 1: section 5)
* weaker than covalent bonds
* between hydrogen and really electronegative atom(O, N, F)
* allows for special properties of water
* between hydrogen and really electronegative atom(O, N, F)
* allows for special properties of water
34
New cards
van der Waals attraction(week 1: section 5)
* weakest force of attraction
* nonspecific interaction, can happen in all types of molecules
* nonspecific interaction, can happen in all types of molecules
35
New cards
Hydrophobic force(week 1: section 5)
* similar types of forces interacting w/ e/o(hydrophobic w/ hydrophobic)
* helps to promote molecular interactions
* important for building cell membrane
* helps to promote molecular interactions
* important for building cell membrane
36
New cards
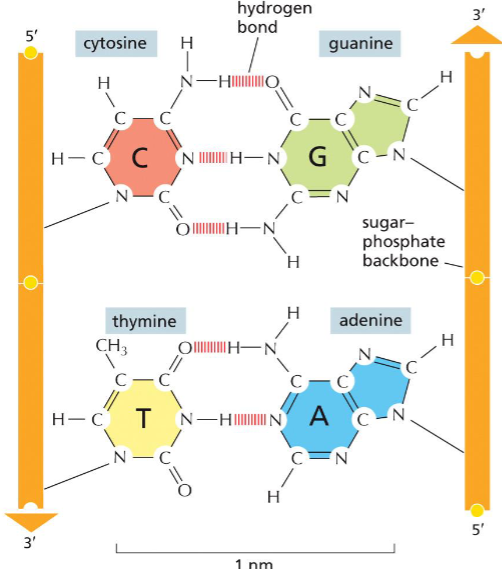
Base pairing(week 1: section 5)
* holds DNA double helix shape together
* A - T has 2 hydrogen bonds
* G - C has 3 hydrogen bonds
* A - T has 2 hydrogen bonds
* G - C has 3 hydrogen bonds
37
New cards
Forces that keep DNA strands together(week 1: section 5)
1. hydrogen bonds
2. hydrophobic interactions
3. van der Waals attractions
38
New cards
Advantages of DNA structure(week 1: section 5)
* energetically favourable conformation
* proteins can recognize + make contact w/ specific DNA sequences in major + minor grooves
* proteins can recognize + make contact w/ specific DNA sequences in major + minor grooves
39
New cards
DNA strands(week 1: section 5)
* 2 strands are complementary
* can be unzipped
* antiparallel (one strand is 5’→3’, other is 3’→5’)
* end of 5’ made of phosphate group(PO4)
* end of 3’ made of hydroxyl group(OH)
* can be separated by proteins in cell + heat
* can be unzipped
* antiparallel (one strand is 5’→3’, other is 3’→5’)
* end of 5’ made of phosphate group(PO4)
* end of 3’ made of hydroxyl group(OH)
* can be separated by proteins in cell + heat
40
New cards
Advantages of separating DNA strands(week 1: section 5)
important for:
* DNA replication
* RNA synthesis
\
* DNA replication
* RNA synthesis
\
41
New cards
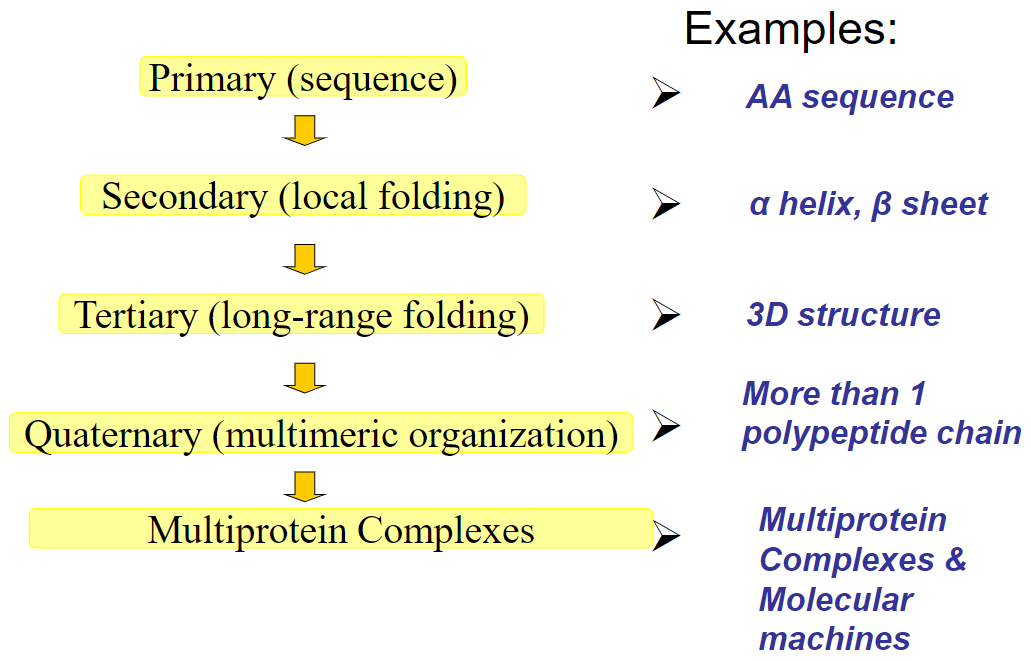
Flow chart of protein structure(week 2: section 2)
42
New cards
Amino acids(week 2: section 3)
* subunits of proteins
* __**Types of amino acids:**__
1. acidic (important for enzymes)
2. basic (important for enzymes)
3. uncharged polar (h-bonds in water)
4. nonpolar (insides of proteins, may be present in lipid bilayers)
* __**Types of amino acids:**__
1. acidic (important for enzymes)
2. basic (important for enzymes)
3. uncharged polar (h-bonds in water)
4. nonpolar (insides of proteins, may be present in lipid bilayers)
43
New cards
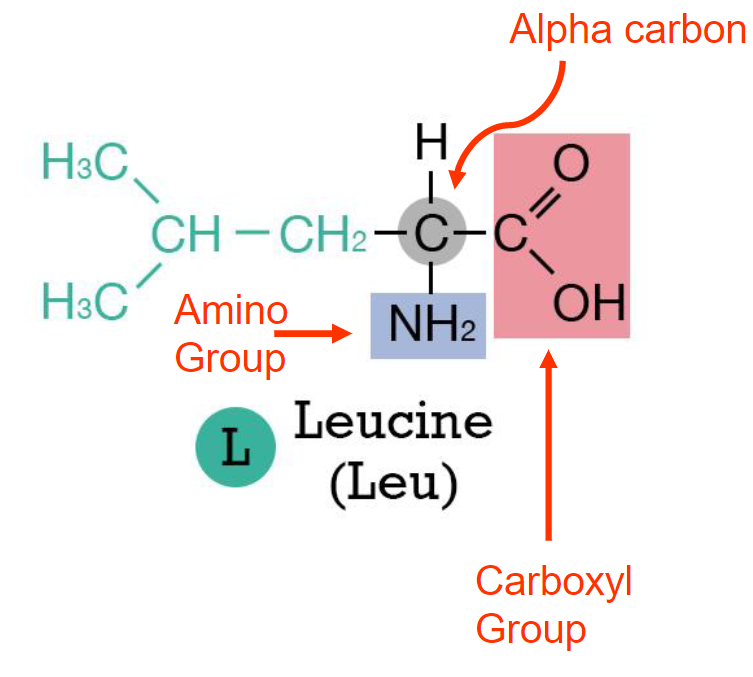
Structure of amino acids(week 2: section 3)
* alpha carbon
* carboxyl group
* amino group
* R group(what decides amino acid)
* carboxyl group
* amino group
* R group(what decides amino acid)
44
New cards
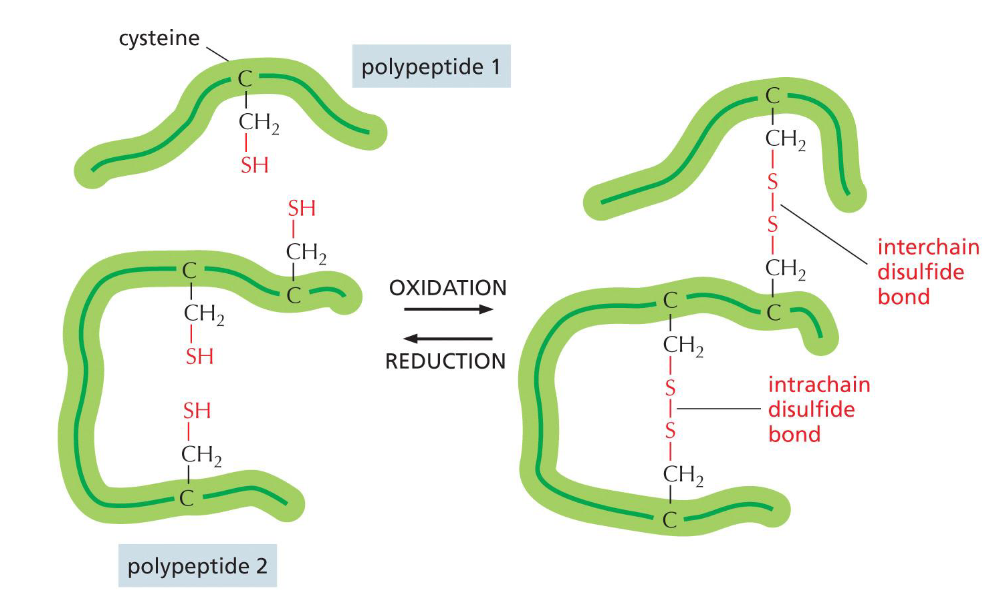
Cysteine’s uniqueness(week 2: section 3)
* has disulfide bonds
* non polar amino acid
* non polar amino acid
45
New cards
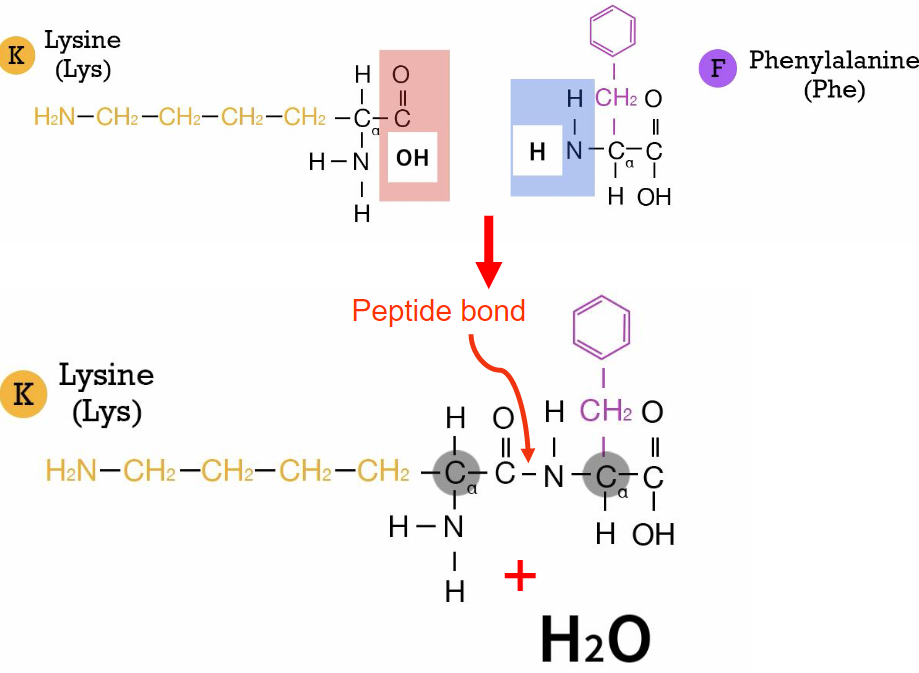
Peptide bonds(week 2: section 4)
* forms btwn carboxyl group + amino group of diff. amino acids
* R groups r not involved
* causes polypeptide chain to have amino end(N terminus) + carbonyl end(C terminus)
* water as product(condensation reaction)
* R groups r not involved
* causes polypeptide chain to have amino end(N terminus) + carbonyl end(C terminus)
* water as product(condensation reaction)
46
New cards
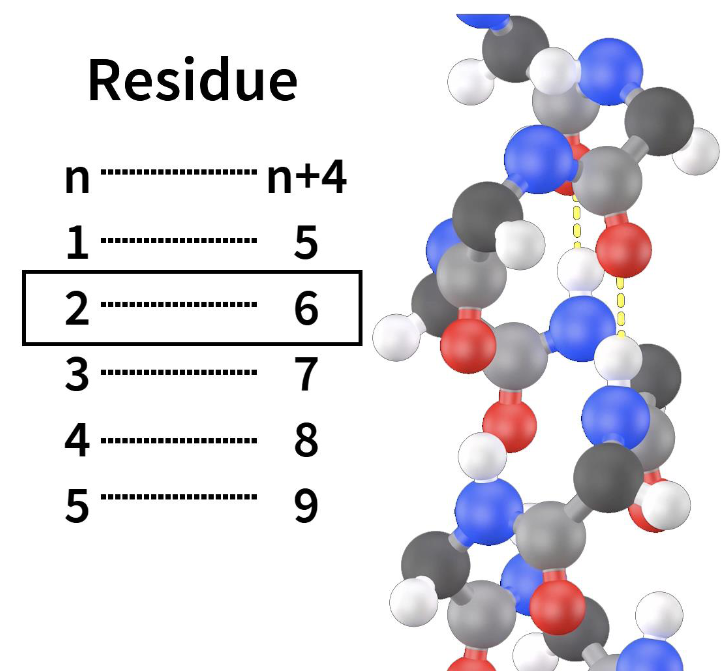
Alpha helix(week 2: section 5)
* has N-terminal + C-terminal
* R groups r not involved
* hydrogen bonds btwn every 4 amino acids(residue)
* R groups r not involved
* hydrogen bonds btwn every 4 amino acids(residue)
47
New cards
Beta sheet(week 2:
* R groups are not involved (but alternately project up + down)
* usually contains 4-5 beta strands, but can have 10+
* H-bonding btwn carbonyl oxygen (C=O) + amine hydrogen (N-H) of 2 diff. amino acids in neighbouring strands
* usually contains 4-5 beta strands, but can have 10+
* H-bonding btwn carbonyl oxygen (C=O) + amine hydrogen (N-H) of 2 diff. amino acids in neighbouring strands
48
New cards
Types of Beta sheets(week 2: section 6)
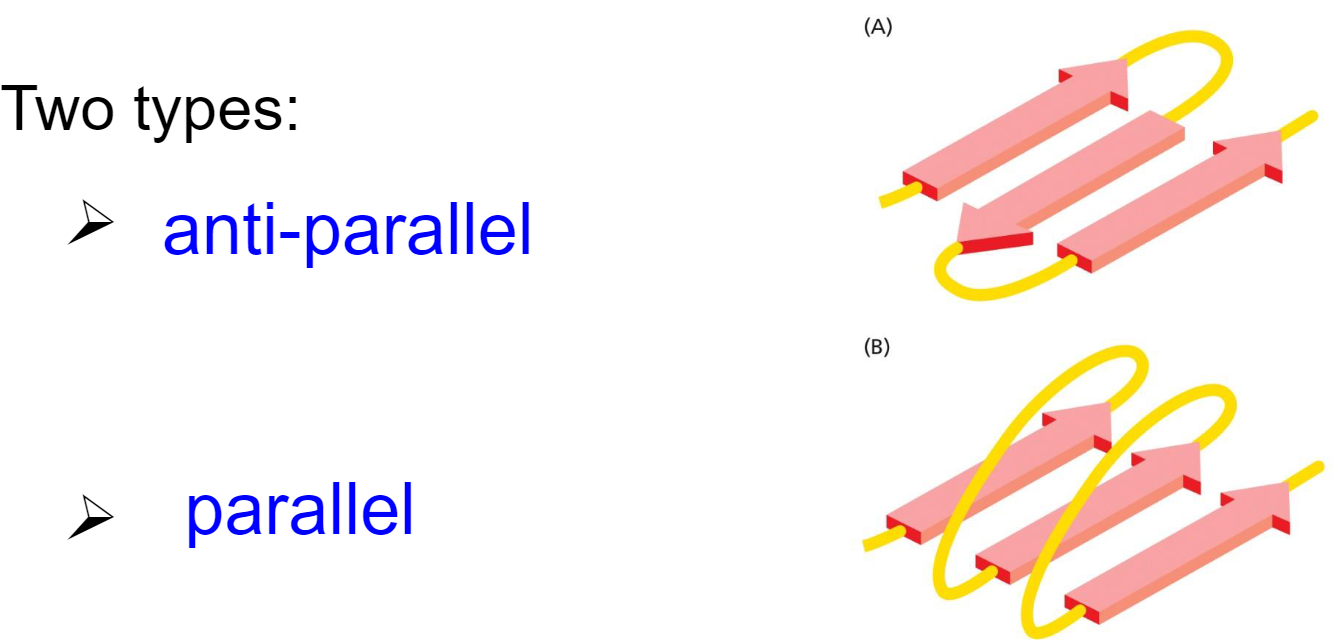
* anti-parallel
* parallel
* parallel
49
New cards
H-Bonding in secondary structures(week 2: section 6)
* __**atoms in bonding:**__ carbonyl oxygen + amine hydrogen in peptide backbone
* __**Alpha helices:**__ h-bonding every 4 AA’s apart within polypeptide chain
* __**Beta sheet:**__ btwn AA’s in diff. segments/strands of polypeptide chain
* __**Alpha helices:**__ h-bonding every 4 AA’s apart within polypeptide chain
* __**Beta sheet:**__ btwn AA’s in diff. segments/strands of polypeptide chain
50
New cards
Coiled coils(week 2: section 6)
* multiple alpha helices tied together
* amphipathic (has both hydrophilic + hydrophobic parts)
* found in alpha-keratin of skin, hair + myosin motor proteins
* amphipathic (has both hydrophilic + hydrophobic parts)
* found in alpha-keratin of skin, hair + myosin motor proteins
51
New cards
Tertiary structure(week 2: section 6)
* overall 3D structure of a protein
* proteins fold into conformation that is ==most energetically favourable==
* protein ==shape dictated by amino acid sequence== aided by chaperone proteins
* __**held together by:**__
1. hydrophobic interactions
2. non-covalent bonds
3. covalent disulfide bonds
* proteins fold into conformation that is ==most energetically favourable==
* protein ==shape dictated by amino acid sequence== aided by chaperone proteins
* __**held together by:**__
1. hydrophobic interactions
2. non-covalent bonds
3. covalent disulfide bonds
52
New cards
Chaperone proteins(week 2: section 6)
* helps the process of protein folding more efficient + reliable
53
New cards
Diff. models of tertiary structures(week 2: section 6)
1. __**backbone model:**__ shows overall organization of polypeptide chain
2. __**ribbon model:**__ shows folding patterns
3. __**wire model:**__ shows R groups’ positions
4. __**space filling model:**__ shows protein surface
54
New cards
Protein domains(week 2: section 6)
* regions of polypeptide chain that are able to independently fold into tertiary structure
* domains specialized for diff functions
* important for evolution of proteins
* domains specialized for diff functions
* important for evolution of proteins
55
New cards
Protein families(week 2: section 6)
* common evolutionary origin
* have similar aa sequences + tertiary structures
* members evolved to have diff functions
* most proteins belong to families w/ similar structural domains
* have similar aa sequences + tertiary structures
* members evolved to have diff functions
* most proteins belong to families w/ similar structural domains
56
New cards
Quaternary structure: hemoglobin(week 2: section 6)
* hemoglobin protein formed from separate subunits: 2 α, 2 β
* ==each subunit = separate polypeptide chain==
* sickle cell anemia caused by mutation in
β subunit
* ==each subunit = separate polypeptide chain==
* sickle cell anemia caused by mutation in
β subunit
57
New cards
Multiprotein complexes + molecular machines(week 2: section 6)
Can be:
* many identical subunits(proteins)
* mixtures of diff proteins + DNA/RNA (more diverse in function w/ diff protein subunits)
* dynamic assemblies of proteins to form molecular machines
* many identical subunits(proteins)
* mixtures of diff proteins + DNA/RNA (more diverse in function w/ diff protein subunits)
* dynamic assemblies of proteins to form molecular machines
58
New cards
Studying proteins(week 2: section 7)
1. purify protein(s) of interest using electrophoresis/chromatography
2. determine amino acid sequence (using mass spectrometry)
3. discover precise 3D structure
* Proteomics(large scale study of proteins)
59
New cards
Genomes(week 3: section 1)
* can come in all sizes(size not always correlated w/ # of genes/organism complexity)
* includes all DNA including non-coding regions
* includes all DNA including non-coding regions
60
New cards
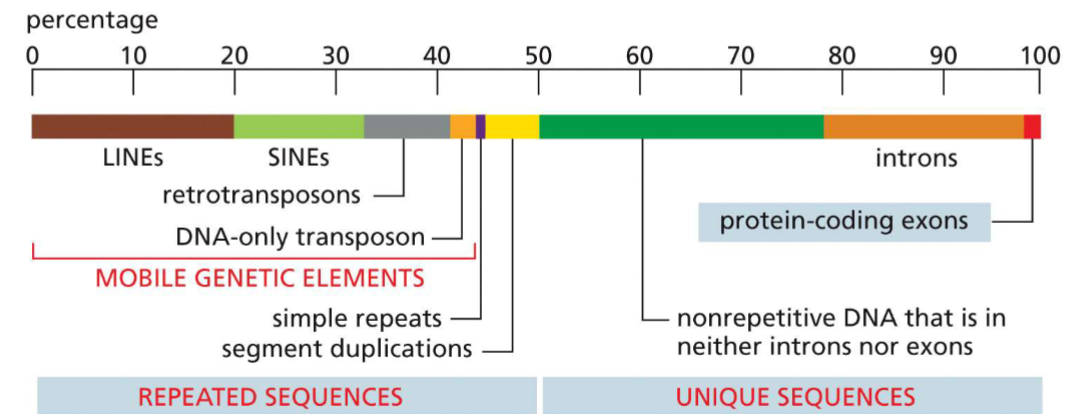
Elements of human genome(week 3: section 1)
__**Repeated sequences(~50%):**__
* simple repeats
* segment duplications
* mobile genetic elements:
1. LINEs
2. SINEs
3. retrotransposon
4. DNA-only transposon
__**Unique sequences(~50%):**__
* nonrepetitive DNA(neither introns/exons)
* introns (transcribed, not translated)
* exons (codes for proteins) (\~1.5% of genome)
* simple repeats
* segment duplications
* mobile genetic elements:
1. LINEs
2. SINEs
3. retrotransposon
4. DNA-only transposon
__**Unique sequences(~50%):**__
* nonrepetitive DNA(neither introns/exons)
* introns (transcribed, not translated)
* exons (codes for proteins) (\~1.5% of genome)
61
New cards
Packing of DNA in the cell(week 3: section 2)
* DNA condensed through folding + twisting, complexed w/ proteins __(genome very big w/o packing)__
* forms the prokaryotic nucleoid
* forms the prokaryotic nucleoid
62
New cards
Eukaryotic genome packing in cells(week 3: section 3)
Challenge:
* human genome very very big**(no personality, smh)**
Solution:
* packing DNA into chromosomes
* human genome very very big**(no personality, smh)**
Solution:
* packing DNA into chromosomes
63
New cards
Fluorescence In Situ Hybridization(FISH)(week 3: section 3)
* uses idea of complementary strands + able to unzip strands
* looks for particular sequence in chromosome(DNA probe __hybridizes__ with chromosome DNA)
* looks for particular sequence in chromosome(DNA probe __hybridizes__ with chromosome DNA)
64
New cards
Chromosomes(week 3: section 3)
* 23 pairs in humans(last pair for sex of human)
* made of chromatin
* replicated in interphase + M phase
* held together at centromere
* ends are called telomeres
* made of chromatin
* replicated in interphase + M phase
* held together at centromere
* ends are called telomeres
65
New cards
Chromatin(week 3: section 3)
* single, long, linear DNA molecule + associated proteins
* tightly packaged but remains assessible for transcription, replication, + repair
* is DYNAMIC(on how tightly packed it is)
* made of 8 different nucleosomes
* tightly packaged but remains assessible for transcription, replication, + repair
* is DYNAMIC(on how tightly packed it is)
* made of 8 different nucleosomes
66
New cards
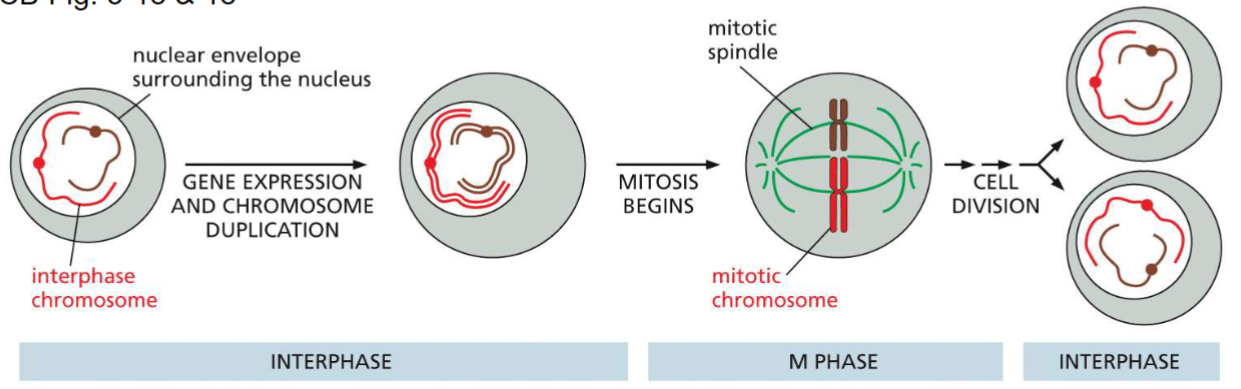
Cell cycle: chromosome replication(week 3: section 3)
__**2 phases:**__
* interphase
* M phase(mitosis)
__**Interphase:**__
* gene expression + chromosome duplication
__**M phase:**__
* mitosis
* chromosome separated
* interphase
* M phase(mitosis)
__**Interphase:**__
* gene expression + chromosome duplication
__**M phase:**__
* mitosis
* chromosome separated
67
New cards
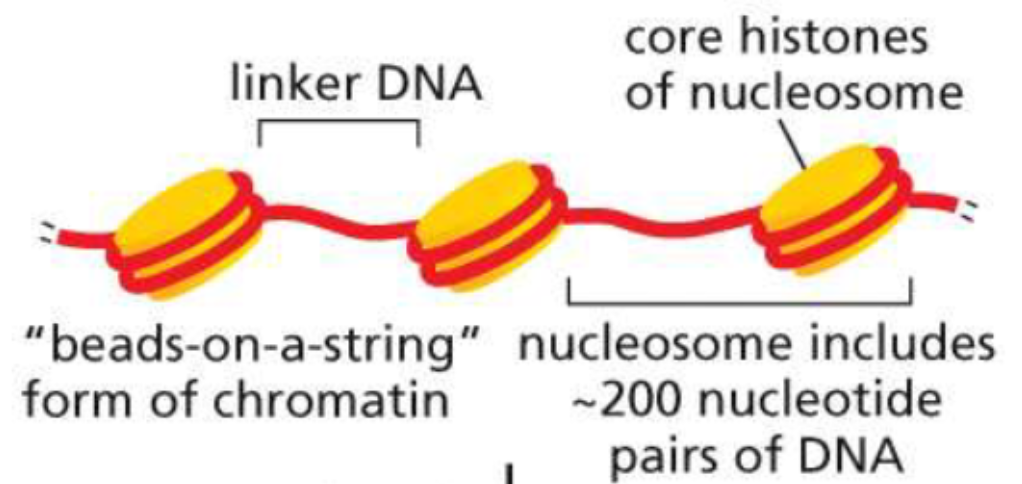
Structure of a nucleosome(week 3: section 4)
* made of DNA wrapped around histones
* \~6 packed histones make 1 nucleosome
* \~6 packed histones make 1 nucleosome
68
New cards
Histones(week 3: section 4)
* small proteins rich in lysine + arginine
* positive charge able to neutralize negative charge of DNA
* 4 core histone proteins:
1. H2A
2. H2B
3. H3
4. H4
* pair of each in octamer core
* 1 linker histone(H1)
* positive charge able to neutralize negative charge of DNA
* 4 core histone proteins:
1. H2A
2. H2B
3. H3
4. H4
* pair of each in octamer core
* 1 linker histone(H1)
69
New cards
Packing of nucleosomes(week 3: section 4)
* non-histone clamp proteins involved in forming chromatin loops
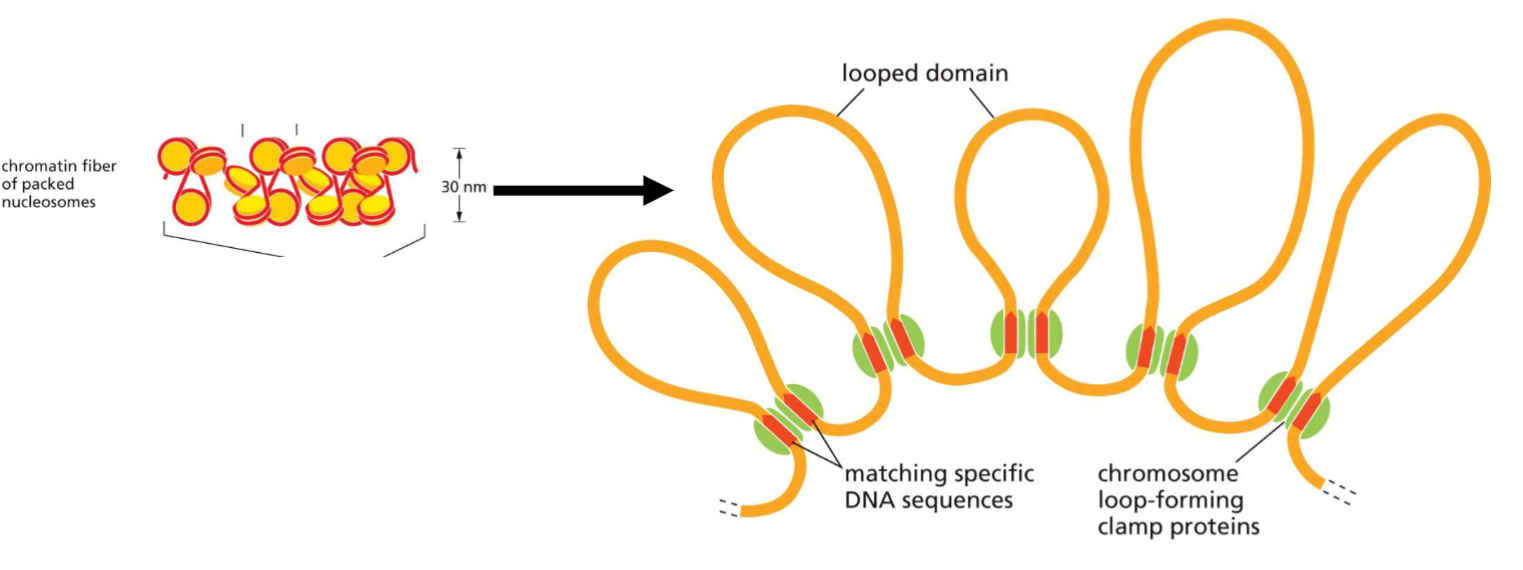
70
New cards
Chromatin packing + re-modeling(week 3: section 5)
__**performed by:**__
* chromatin remodeling complexes
* histone modifying enzymes
__**Can cause:**__
* heterochromatin
* euchromatin
* chromatin remodeling complexes
* histone modifying enzymes
__**Can cause:**__
* heterochromatin
* euchromatin
71
New cards
Heterochromatin(week 3: section 5)
* __**Highly condensed chromatin**__
* areas where gene expression is __suppressed__
__**examples:**__
* meiotic + mitotic chromosomes
* centromeres + telomeres
* one X chromosome in females(Barr body)
* areas where gene expression is __suppressed__
__**examples:**__
* meiotic + mitotic chromosomes
* centromeres + telomeres
* one X chromosome in females(Barr body)
72
New cards
Euchromatin(week 3: section 5)
* relatively non-condensed chromatin
* areas where genes tend to be __expressed__
* areas where genes tend to be __expressed__
73
New cards
Conservatism of DNA replication(week 3: section 6)
* DNA synthesis is __semiconservative__(only one seen in nature so far)
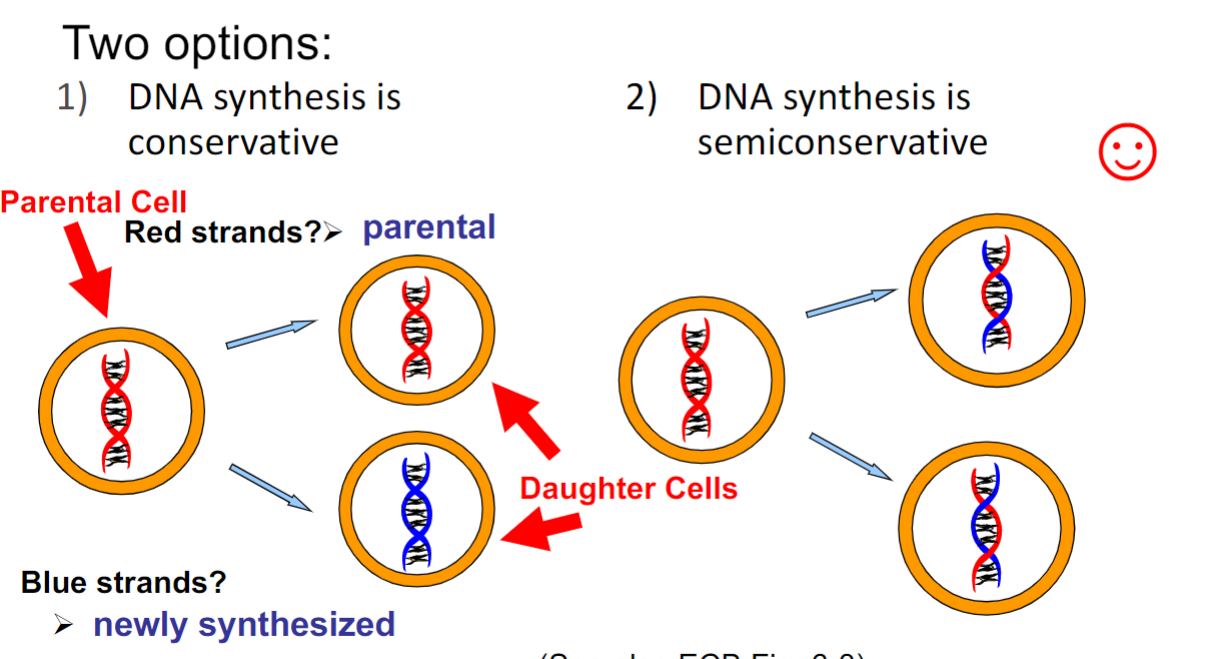
74
New cards
Directionality of DNA replication(week 3: section 6)
* **Always occurs from 3’ end to 5’ end(DNA polymerase stitching)**
* **Growth occurs from 5’ end to 3’ end**
__**3 possible models:**__
1. unidirectional growth of single strands from 2 starting points
2. unidirectional growth of 2 strands from 1 starting point
3. bidirectional growth from 1 starting point
* **Growth occurs from 5’ end to 3’ end**
__**3 possible models:**__
1. unidirectional growth of single strands from 2 starting points
2. unidirectional growth of 2 strands from 1 starting point
3. bidirectional growth from 1 starting point
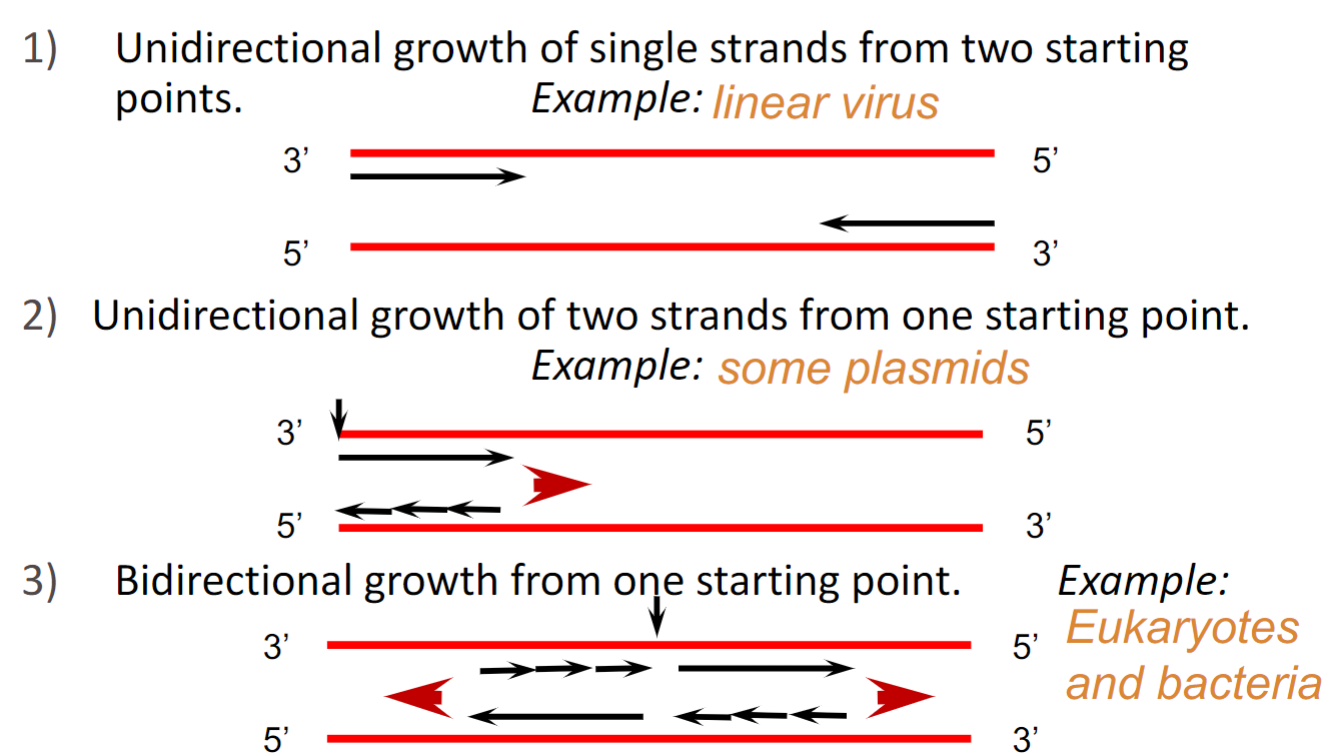
75
New cards
Replication origin(week 3: section 6)
* **Where DNA replication begins**
__**Characteristics:**__
* easy to open, rich in A-T bonds(less h-bonds)
* recognized by and binding of initiator proteins occurs
__**# of origins of replications:**__
* 1 in bacteria
* multiple in Eukaryotes
__**Characteristics:**__
* easy to open, rich in A-T bonds(less h-bonds)
* recognized by and binding of initiator proteins occurs
__**# of origins of replications:**__
* 1 in bacteria
* multiple in Eukaryotes
76
New cards
DNA replication in bacteria(week 3: section 6)
* bidirectional growth from 1 starting point
* this style of replication only applies to circular genomes
* this style of replication only applies to circular genomes
77
New cards
Replication forks(week 3: section 6)
* is asymmetrical
Causes:
* 2 strands
1. __**lagging strand:**__ replicated discontinuously(causes Okazaki fragments)
2. __**leading strand:**__ replicated continuously
Causes:
* 2 strands
1. __**lagging strand:**__ replicated discontinuously(causes Okazaki fragments)
2. __**leading strand:**__ replicated continuously
78
New cards
Initiator proteins for replication(week 4: section 1:
1. binds to origin
2. helps helicase bind
3. requires ATP
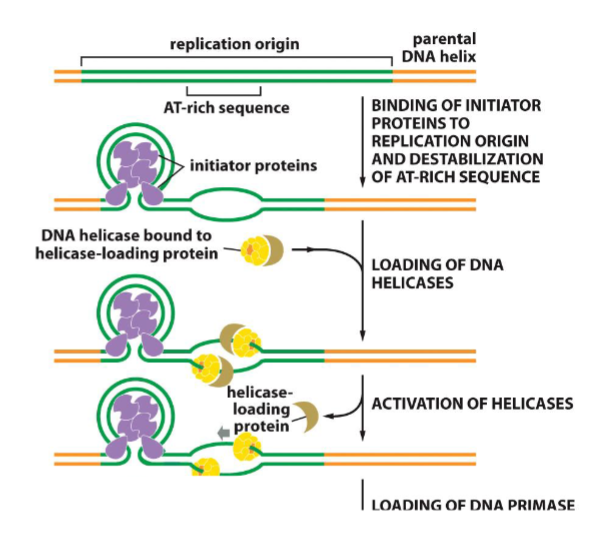
79
New cards
Unwinding DNA(week 4: section 1)
__**Performed by:**__
* 2 types of helicases
* __predominant one__ moves along ==lagging strand== template(5’→3’)
__**Requires:**__
* a lot of ATP
* 2 types of helicases
* __predominant one__ moves along ==lagging strand== template(5’→3’)
__**Requires:**__
* a lot of ATP
80
New cards
Single strand binding proteins(week 4: section 1)
* binds single stranded DNA(ssDNA) to separate strands
* prevents strands from H-bonding, reannealing, hair pins, and loops until replication occurs
* prevents strands from H-bonding, reannealing, hair pins, and loops until replication occurs
81
New cards
Primase(week 4: section 1)
* synthesize RNA primers needed for DNA polymerase to bind
* proceeds(reads) in 3’→5’ along template strand
* proceeds(reads) in 3’→5’ along template strand
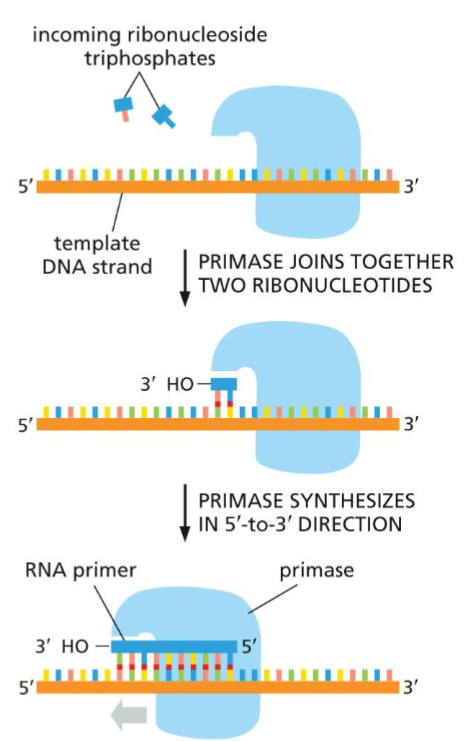
82
New cards
DNA polymerase(week 4: section 1)
* reads 3’→5’ along parent strand
* creates DNA in 5’→3’ direction
* removes 2 phosphates from nucleoside triphosphate to add onto growing strand
* creates DNA in 5’→3’ direction
* removes 2 phosphates from nucleoside triphosphate to add onto growing strand
83
New cards
Sliding clamp(week 4: section 1)
* holds polymerase onto DNA
84
New cards
DNA ligase(week 4: section 1)
* seals nick(gap) caused by removal of RNA primers
85
New cards
Primosome (week 4: section 1)
* helicase + primase
86
New cards
Unwinding problem(week 4: section 2)
Problem:
* as helicase unwinds DNA, supercoiling + torsional strain increases
* problem in circular chromosomes + large linear eukaryotic chromosomes
Solution:
* solved by DNA topoisomerase (breaks phosphodiester bond and reseals it)
* as helicase unwinds DNA, supercoiling + torsional strain increases
* problem in circular chromosomes + large linear eukaryotic chromosomes
Solution:
* solved by DNA topoisomerase (breaks phosphodiester bond and reseals it)
87
New cards
Loss of DNA problem(week 4: section 2)
__**Problem:**__
* major problem for __lagging strand__
* loss of sequence information on 5’ end on daughter strand
__**Solution:**__
* repetitive sequence added to the 3’ end of parent strand determined by RNA template in telomerase
* major problem for __lagging strand__
* loss of sequence information on 5’ end on daughter strand
__**Solution:**__
* repetitive sequence added to the 3’ end of parent strand determined by RNA template in telomerase
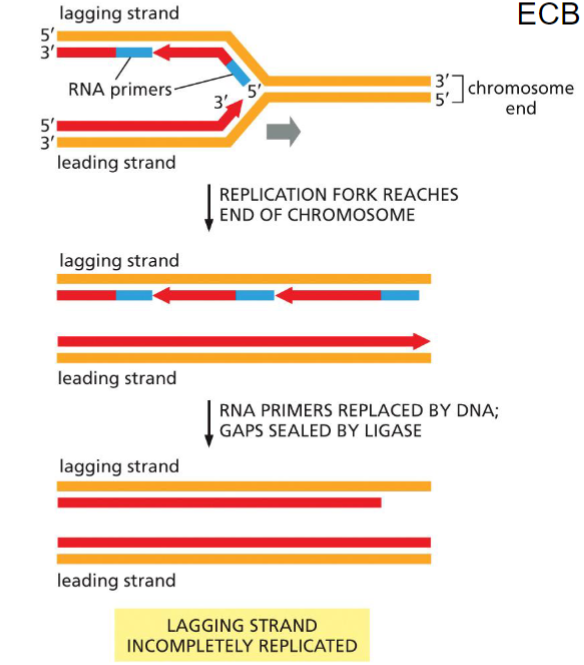
88
New cards
Telomere replication(week 4: section 2)
* RNA template
* resembles reverse transcriptase
* generates G-rich ends
* adds nucleotides to 3’ ends to parental strand template
* resembles reverse transcriptase
* generates G-rich ends
* adds nucleotides to 3’ ends to parental strand template
89
New cards
Telomeres and cancer(week 4: section 2)
* __**telomerase**__ are abundant in stem and germ-line cells, __but not in somatic cells__
* loss of telomeres during DNA replication, limits # of time cell can divide
* Most cancer cells produce high level of telomerase
* loss of telomeres during DNA replication, limits # of time cell can divide
* Most cancer cells produce high level of telomerase
90
New cards
Issues in DNA replication(week 4: section 2)
* if mistake during replication not repaired, mutation occurs and stays in new generations
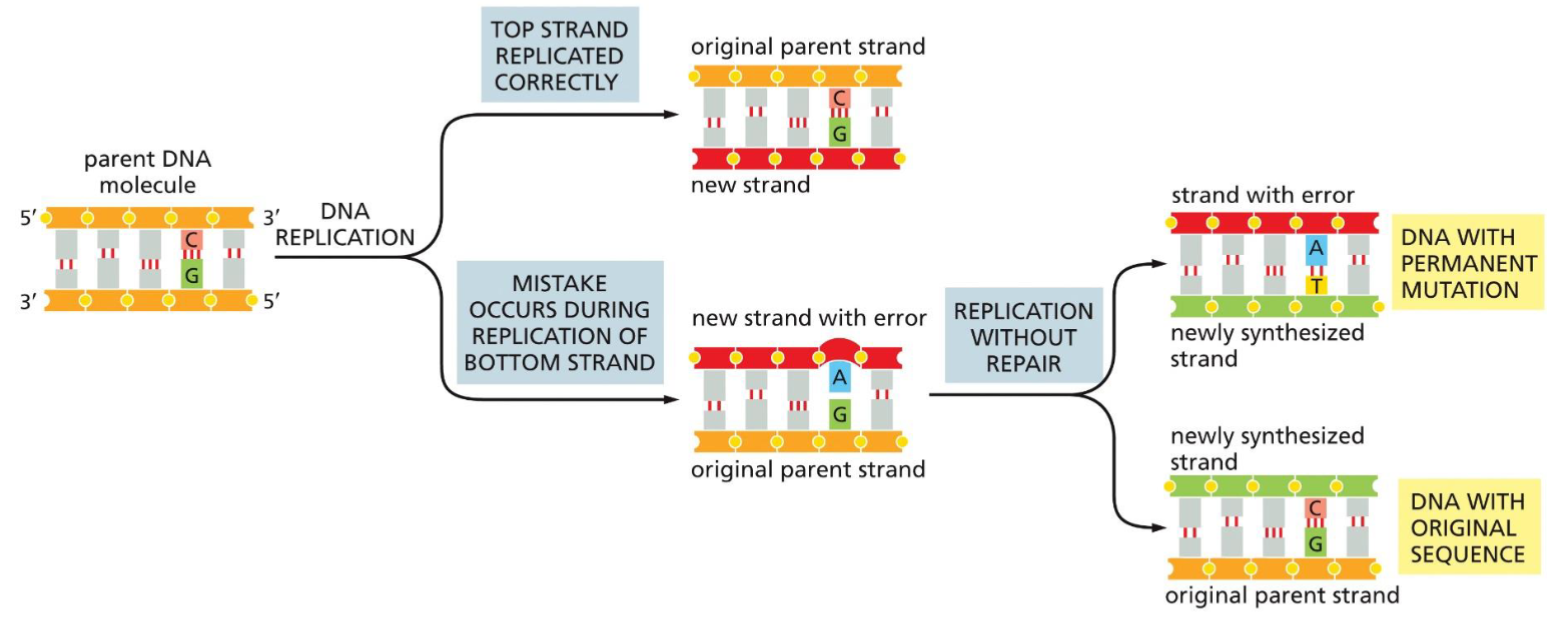
91
New cards
High fidelity of DNA replication(week 4: section 2)
__**RNA polymerases:**__
* has error rate \~1 in 1000
__**DNA polymerases:**__
* has error rate \~1 in 1000000000
\
* human genome(3 bill. bp) only changes \~3 nucleotides every time a cell divides
* has error rate \~1 in 1000
__**DNA polymerases:**__
* has error rate \~1 in 1000000000
\
* human genome(3 bill. bp) only changes \~3 nucleotides every time a cell divides
92
New cards
DNA proofreading + repair: 3’ to 5’ exonuclease(week 4: section 2)
__**Function:**__
* removes misincorporated nucleotide
* performed by DNA polymerase(polymerizing section(P) + editing section(E)
* DNA pol. detects helix distortion and moves back 1 space to remove nucleotide
* removes misincorporated nucleotide
* performed by DNA polymerase(polymerizing section(P) + editing section(E)
* DNA pol. detects helix distortion and moves back 1 space to remove nucleotide
93
New cards
DNA proofreading + repair: strand-directed mismatch repair(week 4: section 2)
* error repair process(when proofreading fails)
* initiated by direction of distortion in geometry of double helix generated by mismatched base pairs
* initiated by direction of distortion in geometry of double helix generated by mismatched base pairs
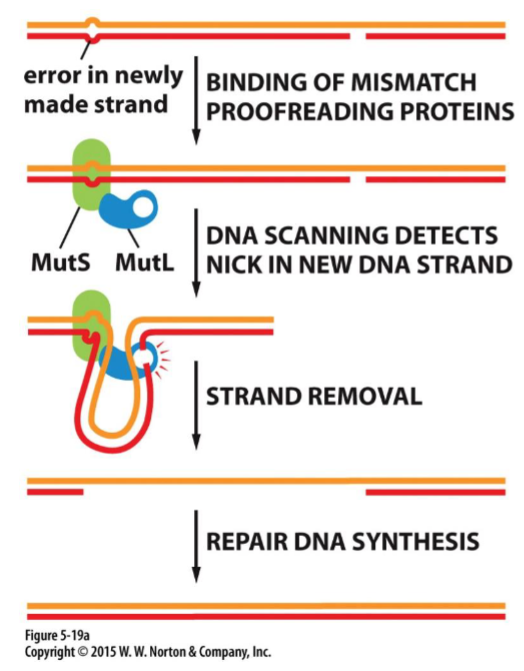
94
New cards
DNA damage(week 4: section 2)
* even after synthesis, DNA can get damaged + need repair
* defects in repair mechs., linked w/ variety of human diseases
__**Types of damage:**__
1. oxidation
2. radiation
3. heat
4. chemicals
* and other cell stressors
* defects in repair mechs., linked w/ variety of human diseases
__**Types of damage:**__
1. oxidation
2. radiation
3. heat
4. chemicals
* and other cell stressors
95
New cards
Spontaneous damage to DNA(week 4: section 2)
__**Depurination:**__
* loss of purines(A,G) in nucleotide
* causes deletion mutation
__**Deamination:**__
* loss of amine(NH2) group on cytosine(C)
* converts C to U
* improper base pairing mutation
* loss of purines(A,G) in nucleotide
* causes deletion mutation
__**Deamination:**__
* loss of amine(NH2) group on cytosine(C)
* converts C to U
* improper base pairing mutation
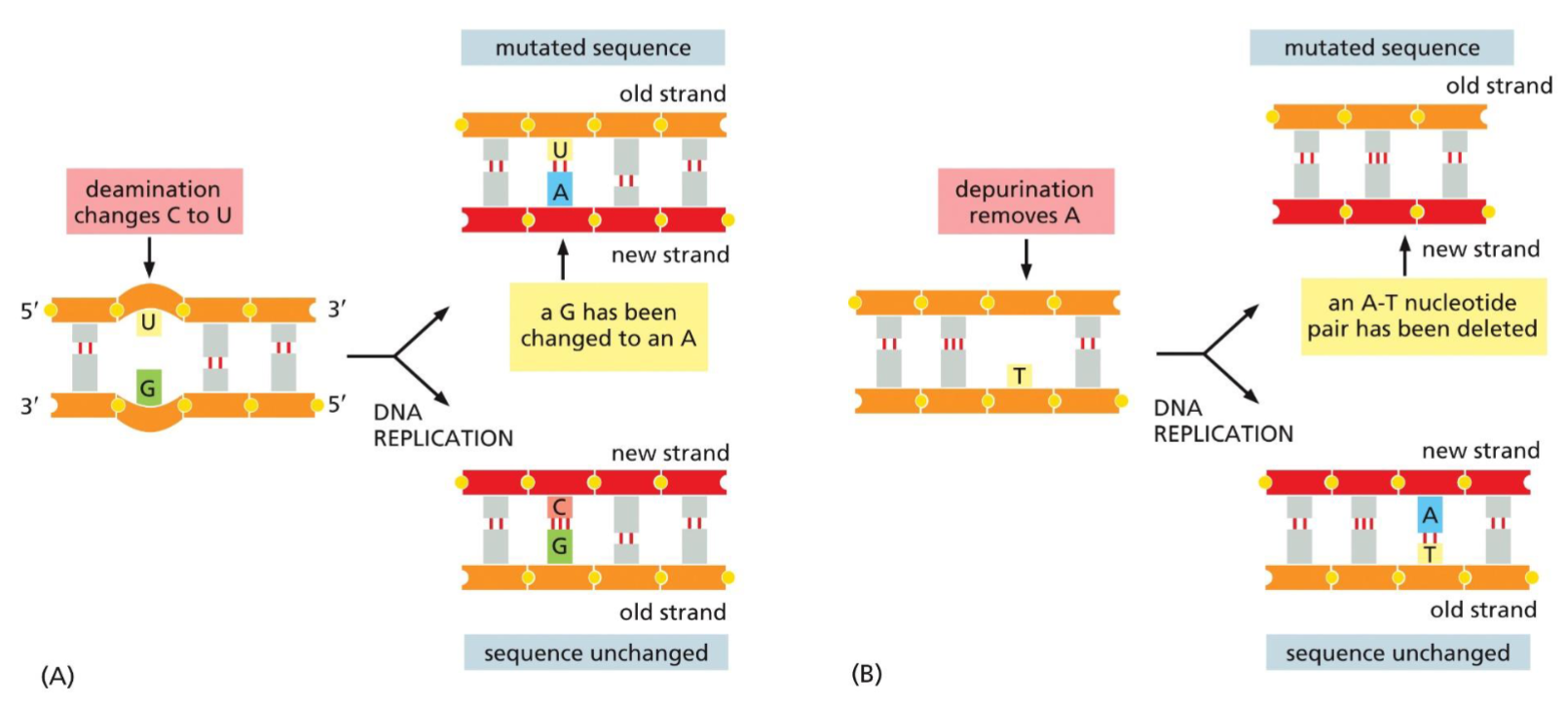
96
New cards
DNA repair mechanisms(week 4: section 2)
1. __**base excision repair:**__ fixes smaller problems(1 base removed)
2. __**nucleotide excision repair:**__ removes multiple nucleotides(ex: dimers)
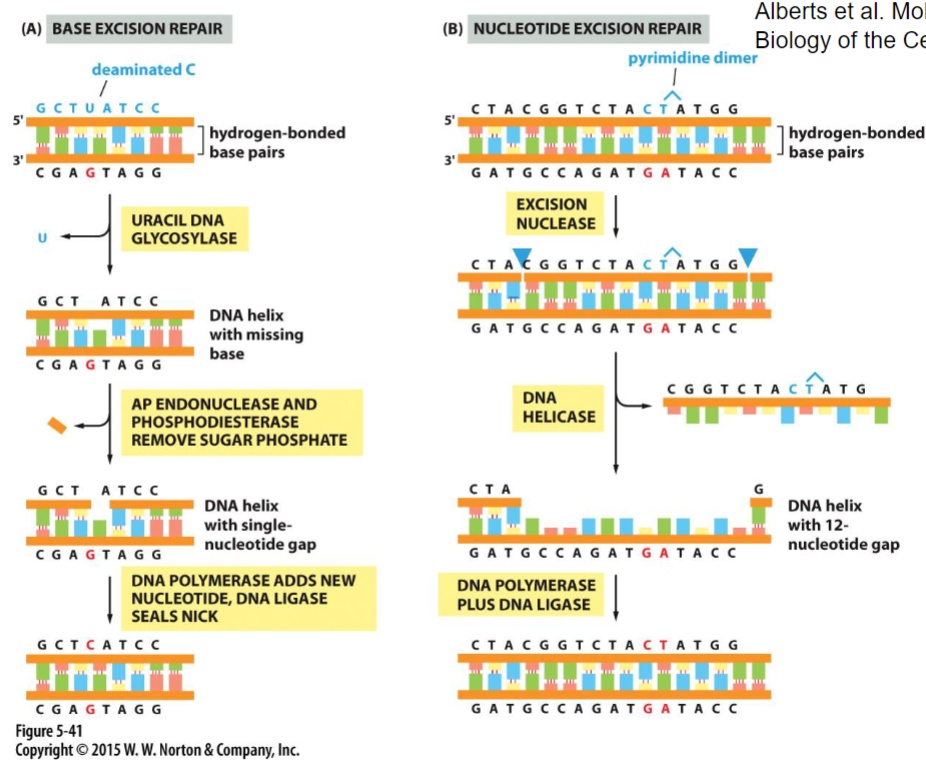
97
New cards
DNA repair of double-stranded breaks(week 4: section 2)
__**Two situations:**__
1. __nonhomologous end joining:__ results in ==some== ==loss of nucleotides at repair site==
2. __homologous end joining:__ results in ==no loss of nucleotides at repair site==
1. __nonhomologous end joining:__ results in ==some== ==loss of nucleotides at repair site==
2. __homologous end joining:__ results in ==no loss of nucleotides at repair site==
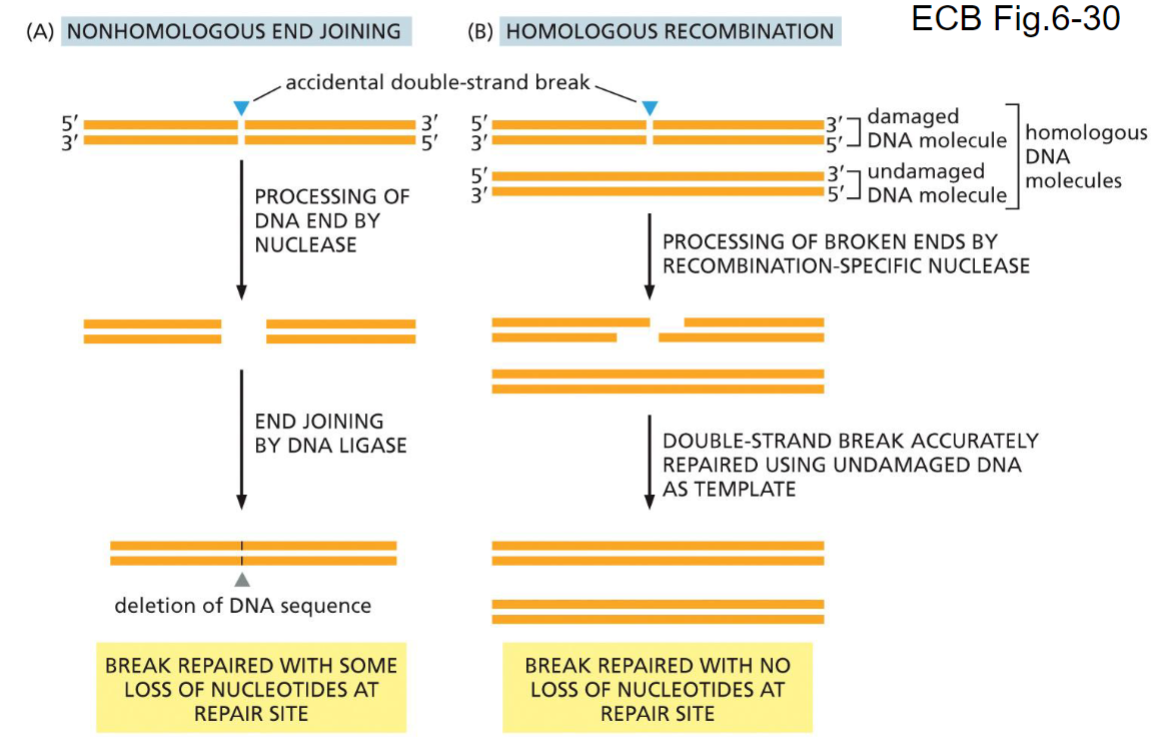
98
New cards
Molecular definition of a gene(week 5: section 1)
* Segments of DNA that are transcribed into RNA
* __**Types of genes when transcribed:**__
1. RNA that encodes for a protein(mRNA)
2. RNA that functions as RNA and may not be translated into protein(tRNA + rRNA)
* __**Types of genes when transcribed:**__
1. RNA that encodes for a protein(mRNA)
2. RNA that functions as RNA and may not be translated into protein(tRNA + rRNA)
99
New cards
Generation of RNA transcript(week 5: section 2)
* RNA nucleotides added in 5’→3’ (anti-parallel)
* uses ssDNA as __template__(other ssDNA is coding strand)
* RNA nucleotides linked by __phosphodiester bonds__
* DNA-RNA helix held by __base pairing__
* uses ssDNA as __template__(other ssDNA is coding strand)
* RNA nucleotides linked by __phosphodiester bonds__
* DNA-RNA helix held by __base pairing__
100
New cards
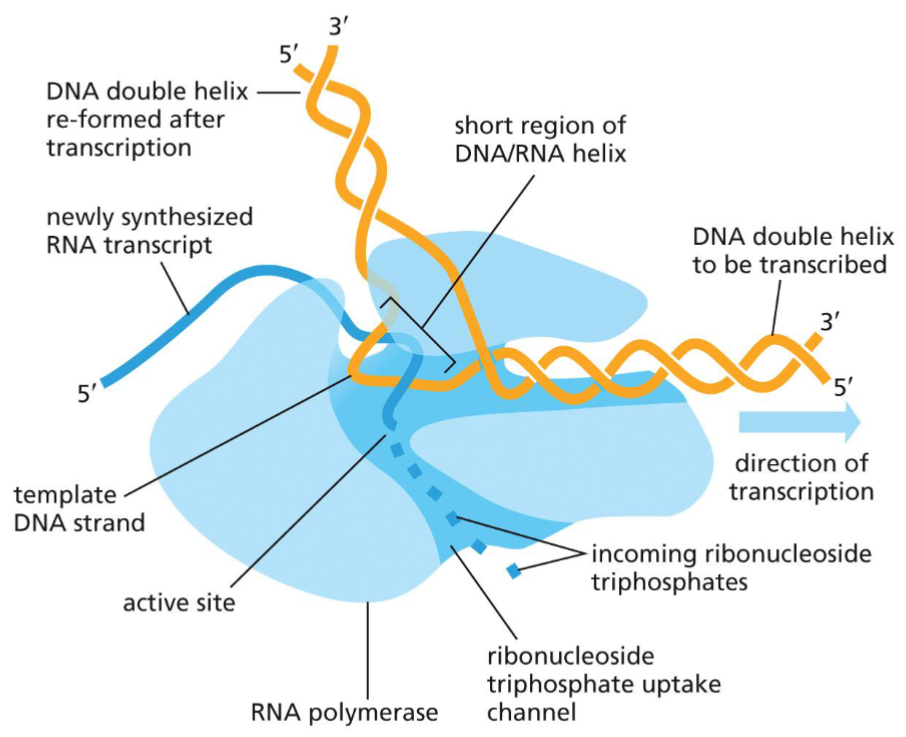
Schematic of RNA polymerase(week 5: section 2)
* no need for primers
* just needs the temple
* less accurate than DNA pol.(more mistakes)
* just needs the temple
* less accurate than DNA pol.(more mistakes)