BIOL 325 LECTURE 13 MOLECULAR DETECTION OF INHERITED DISORDERS
1/94
Earn XP
Description and Tags
Exam 4
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
95 Terms
what is compound heterozygous?
2 mutant alleles but the alleles aren’t the same
alleles are mutated in different locations, really changing the phenotype
what is incomplete dominance or reduced penetrance?
presence of mutation but no abnormal or a range of weak phenotypes
have the mutation but the phenotype can be intermediate or in between or range of phenotypes
what is variable expressivity (SEVERITY)?
range of phenotypes from the same genetic mutation
the range of phenotypes is greater than that of reduced penetrance and more severe with just one mutated allele
not always severe, huge range
associated with >30 monogenic disorders
there are more than 2 different cell types in one individual so COLD-PCR is used for co-amplification
what is genetic heterogeneity? what is locus level and allelic level?
different mutations cause the same phenotype
locus level (multiple genes) = often observed in diseases with multiple genetic components like hemochromatosis
allelic level (one gene) = uncharacterized alleles can still cause a phenotype like cystic fibrosis, muscular dystrophy, and ALS which all have many mutant alleles that can affect phenotype
what is gonadal mosaicism?
somatic mutation in germline cells (gonads)
affects the individual, not the offspring
what is genomic imprinting? what are the three ways that affect the gene or chromosome in genomic imprinting? how are nucleotides and histones involved?
a change in gene expression due to something that happened during the production of the egg or the sperm/gametes
three ways:
gene silencing by turning off a gene or silence a gene by specifically methylating that region in the chromosome
females have a silenced X chromosome and a gene only expressed in some situations (autosome) like if you have brown eyes not blue
large chromosomal deletion
uniparental disomy
one parent gives two chromosomes when the other gives none
nucleotide or histone modifications that do not change the DNA sequence but affect phenotype
NT and histones affect the expression of DNA and environment
what is nucleotide repeat expansion?
increased allele sizes disrupt gene expression and is very commonly tested for
what is mitochondrial inheritance? what is homoplasmy and heteroplasmy? what does the severity depend on?
maternal inheritance
homoplasmy = all mitochondrial DNA in a cell is the same
heteroplasmy = some mitochondria are normal, others have mutations
in one cell, it can have mitochondria with different genotypes/array of phenotypes and is hard to diagnose
the severity of the disease phenotype depends on the amount of mutant and normal mitochondria present
example of incomplete penetrance
what are the four types of genetic etiology?
autosomal vs sex linked
recessive vs dominant (or incomplete dominant)
polygenic
less than complete penetrance
what are the three types of epigenetic etiology? (on top of genes, inherited, and environmental)
genomic imprinting = inactivates one allele/X chromosome (X-inactivation)
inactivates a bunch of genes
methylation of C in CpG islands mostly found in the promoter
allows to turn the gene on and off
chromatin remodeling (binding proteins and histone acetylation)
change the shape of chromatin and degree of compaction which can change its expression
what is multifactorial etiology?
polygenic (many genes) and the environment can cause disease or condition (polygenic ± somatic)
the environment plays a big role along with genetics
what is faster than karyotyping?
molecular detection can be cheaper/faster and karyotyping is very expensive
what does molecular detection detect?
translocation, inversion, duplication, deletion, marker chromosome
what is lethal in humans but is ok in plants?
trisomy
what is mosicism?
forms a chimera
spattering of diff genotypes and phenotypes and you might need to look at multiple cells
aberration must be 4×106 base pairs to detect by karyotype
smaller irregularities are detected by FISH or microarray
what is the derivative chromosome nomenclature?
number of chromosomes, sex chromosomes, chromosome # is a derivative chromosome (der(#)), the derivative of chromosome (der(#)t), then normal chromosome naming
what are marker chromosomes? what is the chromosome that tends to get messed up?
a rearranged chromosome whose genetic origin is unknown based on its G-banded chromosome morphology and is usually present in addition to the normal chromosome complement
it is a typically a supernumerary (more than 46) chromosome and can be composed of inactive genetic material with little or no effect, or can carry active genes and can cause genetic conditions
chromosome 15 tends to be the most common chromosome that gets beat up into different arrangements and a third one is often thrown in
what are single gene disorders?
disease caused by one genetic mutation meaning one gene has been mutated
what are sex-linked genes?
usually on X (Y is too small) and males are hemizygous meaning that males are most likely to express a sex-linked autosomal recessive trait
anything on a sex chromosome means you have to express it with no backup plan
what assumes prediction of a transmission pattern?
mendelian inheritance
mendelian follows the dominant-recessive which doesn’t happen very often, but does happen
what is there no such thing in mendelian inheritance?
incomplete penetrance
variable expressivity
polygenic inheritance (many genes make one phenotype)
basically is not incompletely penetrant or variably expressing and not polygenic
what is penetrance? what is an example?
frequency of phenotype expression in an individual with a gene mutation and is not always predictable
ex = BRCA 1 → just because you have inherited it doesn’t mean you will get breast cancer and if you did get breast cancer then it may not be as severe but it COULD be severe
what are autosomal recessive disorders (AR)?
it is the most frequently observed disorder
the phenotype observed in the heterozygous (+/-) will not have the disorder
individual inherited one mutant allele and will not express it and to show the phenotype, the mutation must be homozygous/must have two mutant alleles
AR genes on pedigrees can skip generations
what are some examples of AR disorders?
cystic fibrosis (varying penetrance)
sickle cell anemia (heterozygous has phenotype)
tay sach’s disease (intermediate phenotype at biochemical level)
fatal disorder where the fatty acids tend to deposit in their nervous system and coats their neurons (life expectancy is 5)
what are autosomal dominant (AD) inherited disorders?
visible in both homozygous and heterozygous state (expressed phenotype)
only need ONE mutant allele
not inherited more often because natural selection cleanses them and will not allow them to reproduce
you WILL see if an individual has an AD disorder
AD inherited disorders do not skip generations
what are some examples of AD disorders?
neurofibromatosis 1 and 2
huntingtons disease
polycystic kidney disease
how do you read a pedigree chart?
males are square and females are circle
if the squares or circles are filled in that means that they have the disease
a slash through the square or circle means they’re deceased
carriers are grey and everyone are connected through lines with double lines indicating incest
what is a pedigree used to determine?
inheritance patterns (somatic, sex linked, X or Y linked disorders and if it is AD or AR)
how would an X-linked recessive disorder look on a pedigree?
the daughters would be gray since theyre carriers and the males would be colored in since they are infected
the daughters have another x chromosome that balances out the other mutated chromosome so they are not affected but males only have one and have no choice but to express it
recognize how Mendelian (AR, AD) and non-Mendelian patterns of inheritance are exhibited by pedigree diagrams (written response question)
In autosomal dominant disorders, it does not skip generations. In autosomal recessive disorders, it can skip generations. The non-Mendelian X-linked recessive pedigree diagrams show that the father gives the mutated X allele to their daughters/females, making them carriers, and the daughters have a chance of giving the X-linked traits to their sons.
what are X-linked disorders?
females must be homozygous to show the phenotype, while males are hemizygous for the mutant allele and will show the phenotype
one X is inactivated in females resulting in mosaicism (Barr body)
can become unsilences if you need a healthy chromosome
what are Y-linked disorders? what are the disorders that are Y-linked?
males inherit Y from dad
easy to pick up because the dad’s y-chromosome has to be given to the offspring
traits that are linked to Y are hemizygous
very rare
Y-linked disorders
Y-linked deafness
Hairy pinea
Short stature homeobox
what are some of the common and well-known X-linked disorders?
muscular dystrophy
one kind of hemophilia
colorblindness
fragile X mental disorder
what are the five possible genotypes and example them?
X+Y = hemizygous wild type male
XmY = hemizygous mutant male
X+X+ = homozygous wild female
X+Xm = heterozygous female carrier
XmXm = homozygous mutant female
describe the transmission patterns of AR, AD, and X-linked disorders (written response question)
In autosomal recessive disorders, the individual must have both mutated alleles to show the phenotype. Heterozygous individuals will not show the phenotype. AR disorders can also skip generations, unlike AD disorders. In autosomal dominant disorders, the individual will show the phenotype in both a homozygous and a heterozygous state. In X-linked disorders, males are mostly affected as they are hemizygous, while females will be carriers due to being homozygous for the X chromosome. Females can still express the disorder, but it is rare.
what are the sex linked penetrance patterns in females?
it ranges from low to severe
don’t even know you have it until its bad
X chromosome inactivation prevents overexpression of genes contained on the X chromosome in females
the reasons for varying severity are autonomous expression, skewed X-inactivation, clonal expansion, and somatic mosaicism
what are the sex linked penetrance patterns disorders that do not fit standard X linked recessive or X linked dominant rule?
adrenoleukodystrophy and fragile x syndrome
what are the sex linked penetrance patterns in males?
severity and penetrance is generally very high
will still show some range in severity but will be generally close to severe
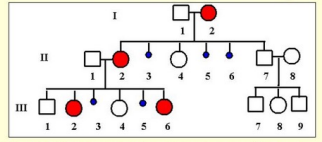
what is the X-linked dominant inheritance with male lethality?
males that have inherited one allele, it is lethal for them
mother has the x-linked mutations and passed it onto her daughters but they are ok since they are homozygous for the x chromosome
if the daughters give birth to a male, male will most likely die
the others on did not die which is an example of odd penetrance
males can have a low phenotype/low penetrance effect so the males CAN survive but it is most likely on the higher end, causing lethality in most males
only females and boys with Klinefelter syndrome are affected
the pedigree may appear to have several spontaneous abortions in the offspring of the affected females
what are the examples of x linked dominant inheritance with male lethality?
incontinentia pigmenti and aicardi syndrome
what is autonomous expression?
expressed later in life and it is severe and is highly expressed in the organism
what is loss of heterozygosity (LOH)?
loss of the normal allele, revealing the mutant allele
commonly happens in both inherited disorders, especially in the cancer realm
autosomal recessive mutations can result in an abnormal phenotype in a hemizygous situation where the mutant allele is the only allele expressed
will not show a phenotype with a wild type/deletion state since the wild type is balancing out the mutant allele
what does LOH result from?
results from somatic (environmental, not inherited) mutations or deletions of the normal allele and often result in cancer
contrast GOF, LOF, and dominant negative mutations (written response question)
gain of function (GOF) mutations usually display a dominant phenotype and only need to have a mutation in only ONE allele to have a GOF mutation. gain of function mutations cause proteins that either increase their activity or lengthen their functional lifespan. loss of function (LOF) mutations usually display a recessive phenotype and are also called haploinsufficiency. LOF mutations can happen if the gene results in expression of only half the normal amount of functional protein, which is insufficient to maintain normal function. dominant negative mutations are observed in loss-of-function mutations of multimeric proteins with quaternary structures, which can be utilized by single or multiple genes. it is often due to mutations that suppress the activity of proteins that are multimeric with themselves or others.
what does haploinsufficiency mean?
one gene may not be enough for the needed function in the organism/one gene is insufficient
what are dominant negative mutations?
very common in single transduction pathways
all the receptors in single transduction signal bind to a receptor, like answering a phone, and the cell communicates information
the receptors in single transduction pathways lead to subunits and what happens is one subunit is wild type and if you’re heterozygous, the other subunit is mutant which is the dominant negative
heterozygous individuals may display an impaired function
maybe 25% of your own receptors is enough for you to survive but maybe its not
the mutant protein affects the activity of every protein complex it is a part of, causing more than a 50% decrease in the activity of that protein
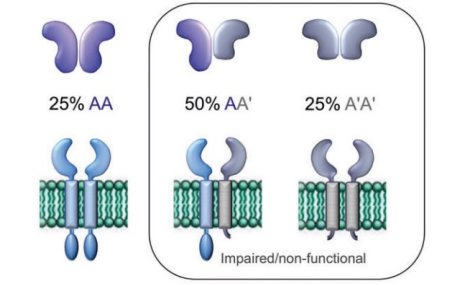
explain this dominant negative figure
25% AA = inherited two normal alleles making multimeric proteins with two identical subunits and functions fine → homozygous wild type
50% AA’ = it is the wild type plus the mutant
don’t know what its going to do and might lead to low functionality or nonfunctionally depending on the gene, safe to say it won’t function well
25% A’A’ = homozygous mutant meaning the receptor will not work
what are antimorphic mutations? whats an example
results in an altered molecular function which is often inactive and are characterized by a dominant phenotype
in humans, dominant negative mutations have been implicated in cancer which are somatic
ex = marfan syndrome which has a DN mutation and haploinsufficiency
how does TP53 exhibit LOH, GOF, and DN mutations?
LOH = wild type isn’t used and mutant is only expressed
DN = combination of two diff forms of that protein being made in the organism
in either situation, you lose the tumor suppressor activity
GOF = p53 mutants might possess new activities, which are not present in the original WT p53 protein
what is the example in class for dominant negative? what are the symptoms? what is the inheritance and what is required for its diagnosis?
vascular ehlers-danlos syndrome = an inherited connective tissue disorder that is caused by defects in the collagen
symptoms are thin, translucent skin, easy bruising, and fragile arteries, muscles, and internal organs
the most severe form of VEDS is caused by a dominant negative mutation in the COL3A1 gene that helps create collagen
collagen has a quaternary structure meaning it needs multiple subunits to hook together
inheritance is autosomal dominant and the effect is commonly dominant negative
inheriting BOTH mutant alleles is worse than inheriting one because you won’t create any normal collagen if you inherit both
diagnosis is tough and almost always requires sequencing because there are a lot of diff alleles and mutations
what is hemochromatosis?
affects men 24x more frequently (variable expressivity)
about 1 in 227 homozygous and 10% are carriers of the worlds population
what is essential for cystic fibrosis?
early ID is crucial for survival and 23 diff mutations require detection
what are monogenic diseases?
one gene/impaired single gene causes the disorder meaning one inherited gene mutation results in the disorder
what is factor v leiden?
most commonly hereditary hypercoagulability disorder
the mutation (R506Q) results in one amino acid substitution which causes thrombosis and is present in the heterozygous form in ~5% of humans
the protein is a multicopper oxidase cofactor for coagulation
autosomal dominant
what does FVL have that varies?
varying PENETRANCE = a lot of people have the mutant vs the wild type but do not have a phenotype so it is not penetrating
varying EXPRESSIVITY = does not mean the severity, it means what is the phenotype
is the phenotype anemia, high blood pressure, etc?
expressivity is how its seen/manifested
how was the FVL mutation detected by PCR-RFLP?
the mutation messes up/destroys a restriction enzyme site
PCR amplifies the DNA so there is enough to evaluate then there are primers that flank the mutation site and RENs are added
better to do PCR before RFLP because the alternative is radiation
a wild type = three bands indicating that the Mn/l site was not destroyed
homozygous mutant = two bands indicating the Mn/l site was destroyed
heterozygous = there will be four bands as it is mutant + wild type
how was PCR + gel electrophoresis used for detecting the FVL mutation?
can pick up FVL mutation by just using primers that flank the mutation site
each subject has two lanes with lanes for a positive control, negative control, and blank
the intensity of the mutation should be the same as the control
those who are positive for the mutation will have a band in their two lanes
how was SSP-PCR used to detect the FVL mutation? when is it used?
PCR amplifies the sample and at the site of the mutation, design a wild type primer and a mutant primer
mutant primer can only anneal to a mutated gene so a longer primer ends on mutated base, making a larger amplicon (148 bp) → one band
wild type primer anneals to the wild-type/normal sequence which makes a shorter amplicon and will show up first (123 bp) → two bands
SSCP can be used if you do not know what is going on and you need to fish around
can also be used if PCR results are different, even if there are signs and symptoms of FVL or if there is a different mutation in a different region
what would the results look like just by using SSCP analysis alone?
homozygous wild type/negative will have bands lower on the gel due to it being smaller
heterozygous mutant will have three bands
homozygous mutant/positive will have two bands higher on the gel due to it being larger
understand how point mutations are detected by molecular methods, such as PCR, PCR-RFLP, SSP-PCR, Invader assays, and Southern blot (written response question)
PCR can be used to help detect molecular methods to directly amplify the target of interest, so there is more to evaluate. With PCR-RFLP, restriction enzymes are used to cut at the specific mutation sites, such as when a site is created or destroyed. SSP-PCR uses specific primers for the wild-type sequence and the mutant sequence, which makes different-sized bands that show up on a gel. Invader assays use probes that form a triplex at the mutation site, causing the cleavage of the arm and generating a fluorescent signal to indicate the presence of the mutation. Southern blot also uses probes to detect point mutations, which can be seen on a gel through radiation.
what is hemachromatosis 1? what are the symptoms and inheritance?
from the overabsorption of iron from food which causes mutations in the gene for a membrane iron transporter (HFE)
transmembrane protein HFE gains function where it absorbs too much iron which can be fatal in children
symptoms = fatigue, joint point, cirrhosis, heart failure, insulin resistance ED, and increased susceptibility to infections
HFE hemochromatosis is AR
the C282Y mutation in 10% of caucasians, disease results in 0.3% of the population which is an example of reduced penetrance
how was the HFE C282Y detected with PCR-RFLP?
a new site (Rsa1) is created due to the mutation
PCR amplifies the whole area, exposing the area that mutates
wild type = has two bands
homozygous mutant = three bands due to new site created
heterozygous = four bands
what is cystic fibrosis? what is the LOF of cystic fibrosis?
commonly caused by a trinucleotide deletion in the CF transmembrane conductance regulator gene but has 1900 other mutations
gene is a hot spot for mutations
causes severe lung damage and nutritional deficiencies
affects cells that produce mucus, sweat, saliva, and digestive juices
supposed to create a normal chlorine channel but it loses its function to create that normal Cl channel
autosomal recessive
what is the ACMG core panel for CF?
detects 23 mutations and will ID 50-98% of carriers but theres a large range due to ethnicity
those in the 50% area will need additional analysis
those in the 98% do not need additional analysis
what are the diagnostic approaches for CF?
RFLP, PCR-RFLP, heteroduplex analysis, SSCR, SSP-PCR, cleavase, bead array, and direct sequencing
what is an example of variable expressivity?
somatic mosaicism is common in neurofibromatosis type 2
mutations in NF2 predispose to nervous system tumors and NF2 is a tumor suppressor gene
what is COLD-PCR?
co-amplifies at lower denaturing temperatures which selectively amplifies minority alleles from mixtures of WT and mutation-containing sequences
COLD-PCR would have similar peaks to standard PCR at 75% or 25% wild type
better at picking up mutations and would have better peaks in tissues that have 1-6% of mutations compared to standard PCR
what is incomplete penetrance?
presence of mutation but no predictable phenotype (could be weak or strong phenotype)
how does cystic fibrosis, muscular dystrophy, and ALS in allele level often lead to variable expressivity or incomplete penetrance? what is the best with these diseases?
cystic fibrosis = patient can have 850 alleles (compound heterozygous) which most genes in the human race don’t have that many alleles
can be heterozygous within a population but heterozygous within yourself meaning there can be a variety of phenotypes
muscular dystrophy = patient can inherit thousands of alleles (X-linked, which is most common in men), affecting the severity of the phenotype
which allele an individual has and if two are combined (which makes it even harder) can affect the phenotype
ALS (amyotrophic lateral sclerosis) = which of the 20+ alleles a patient has can affect the phenotype and even two people with the same phenotype, the disease is different
molecular diagnostics is the best with detecting these diseases as the alleles, the combination of alleles, and the changes of phenotypes can make it hard to diagnose and detect with any array
what is marfan syndrome?
a connective tissue disorder with variable clinical expression
theres also vessels in the connective tissue which means the cardiovascular system is strongly affected by the disorder too
caused by pathogenic variants in FBN1 or other genes AND has overlapping phenotypes with other genetic diseases and differentiating among those diseases is critical for treatment
aortic dysfunction syndromes (marfan, loey dietz, ehlers danlos) are tested by NGS
has a range of phenotypes and different genes that can lead to the disorders and some of them affect the heart more than others and if they affect the heart, they’re serious
the known genes are classified by families as of how you treat it and how severe it is
20 mutations to FBN1 can result in Marfans
how is marfan syndrome detected?
nested PCR
external primers to amplify first then use product to use internal primers
nested multiplex
better and looks for several genes at the same time
want to know which mutation they have so they can be treated and know the severity
single cell expression assays
mosaicism can come into play
discuss why most disorders have complex transmission patterns that have hard to predict phenotypes (written response question)
Most disorders have complex transmission patterns because they are multifactorial. Complex traits have no distinct inheritance patterns and can be affected by multiple loci (genes) and or environmental factors (somatic). The phenotypes of the complex traits are also defined by thresholds of quantitative traits like if an individual is obese or has diabetes.
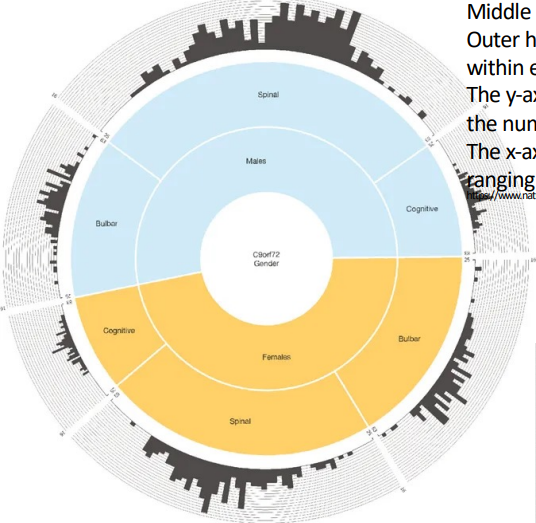
explain the picture for incomplete penetrance of C9orf72 mutants
they all have the same gene mutation but have different phenotypes
incomplete expressivity from no effect to extremely severe effect in the same exact location being mutated
males are more affected than females and the phenotype that is most common between them are spinal
histogram indicates the age (25-60) and how many in that age have the symptoms
name and define five non-mendelian patterns of inheritance that need to be evaluated clinically, as they might require more complex testing. describe an example discussed in class for each (written response question)
Sex-linked inheritance which can be on the X or the tiny Y. Y-linked traits won’t affect females, and X-linked traits will affect males because they are hemizygous. The example for sex-linked inheritance that was discussed in class is hemophilia which is most common in males. Germline/gonadal mosaicism is a somatic mutation in germ-line cells (gonads) and can lead to some offspring being affected while others may not be. An example of germline/gonadal mosaicism that was discussed in class was osteogenesis imperfecta, which is brittle bone disease. Bones break easily and are inherited which can be tested for during pregnancy. Genomic imprinting is a change in gene expression due to something that happened during the production of the egg or the sperm/gametes. Examples of genomic imprinting that were discussed in class were Prader-Wili syndrome which is caused by a deletion or mutation of a paternally inherited chromosome 15 and Angelman Syndrome that is caused by a deletion or mutation of a maternally inherited chromosome 15. Nucleotide repeat expansion is when increased allele sizes disrupt gene functions and the larger the expansion, the greater the disruption in the function of that gene. It is both in the exonic and intronic regions of the genes. An example of nucleotide repeat expansion discussed in class was the hexanucleotide repeat expansion of the ALS mutation. Lastly, mitochondrial inheritance is the maternal inheritance of mitochondrial genes. An example of mitochondrial inheritance that was discussed in class was Kearns-Sayre Syndrome which is a rare neurological disorder that causes progressive paralysis of certain eye muscles, including the eyelid, which can stop functioning.
what are the methods specific for detection of DNA methylation and nucleotide repeat expansion disorders (written response question)(additional to the non-Mendelian patterns)
Four methods could be used for the detection of DNA methylation. Methylation-specific PCR can be used where bisulfite converts unmethylated cytosine to uracil and uses a primer to detect the unmethylated from the methylated. PCR-RFLP with methylation-sensitive restriction enzymes can also be used, where restriction enzymes cut only methylated DNA. Southern blot with methylation-sensitive restriction enzymes can be used as well, and Me-CHIP can be used to look at the whole chromosome to see the degree of methylation using antibodies. The method that is specific for nucleotide repeat expansion is touchdown PCR and repeat-primed PCR. Repeat-primed PCR uses primers outside of the repeat and amplifies the whole thing, which can be used to see their genotype. Repeat primed PCR then uses primers that are three repeats and anneal throughout the PCR reaction, making different-sized PCR products, and creating a sawtooth pattern when the expansion is present. Touchdown-PCR gradually lowers the annealing temperature of a reaction to enhance specific binding of repetitive sequences.
disorder examples and the preferred molecular tests these might be on certification exams (written response question)
Factor V Leiden is often on CLS exams and over three million people in the US have Factor V Leiden. It is a common hereditary hypercoagulability disorder with a mutation in the coagulation factor V. The preferred molecular tests for FVL are PCR-based assays such as PCR-RFLP and SSP-PCR to amplify the target. Hemochromatosis is also often on CLS exams which affects men 24x more frequently, 1 in 227 individuals are homozygous for this mutation, and 10% of the world are carriers. Hemochromatosis is a gain-of-function in the transmembrane protein HFE, where iron is absorbed more, which can be fatal in children. The preferred molecular tests for hemochromatosis are PCR-RFLP and PAGE. Lastly, cystic fibrosis is also commonly found on certification exams and is commonly caused by a trinucleotide deletion. There are a variety of preferred molecular methods for cystic fibrosis, such as RFLP, PCR-RFLP, heteroduplex analysis, SSCR, SSP-PCR, cleavase, bead array, and direct sequencing. There are a lot of diagnostic approaches for cystic fibrosis due to the gene having many mutations.
expand on the non-mendelian inheritance pattern, germline/gonadal mosaicism. what are gonads and what is germline? what would germline mosaicism look like on a gel? what about somatic?
gonads = in the cells of ovaries and testes
germline = specifically the cells that are diploid that are going through meiosis to make the sperm or the egg
there are a lot of genetic tests for diseases and disorders for germline/gonadal mosaicism like X-linked muscular dystrophy and autosomal dominant achondroplasia.
clinically and phenotypically normal parents can have children with a dominant disease
recurrence is possible and testing before second pregnancy is recommended
can be detected by NGS during parental testing
on a gel, the germline mutation would be found in both the blood and sperm meaning it is in multiple tissues not just one, showing that it was likely inherited
in somatic mutations occurring in germline cells, mutation occurred after conception of birth meaning the mutation in an individual developed in their lifetime in their gonadal cells
appears in a single tissue instead of multiple
expand on nucleotide repeat expansion, what is it? what are the repeats? what is the solution in detecting nucleotide repeats? what do the sawtooth patterns indicate?
class of genetic diseases caused by expansions of DNA repeats in both exonic and intronic regions of the gene
can be worse within you but the length of the repeat is inherited
the repeats are STR’s which are never more than 8 repeated units that can expand or contract
the solution in detecting nucleotide repeats are touchdown PCR and repeat-primed PCR
the sawtooth patterns in repeat-primed PCR mean there are more repeats present and you would confirm this by doing the other strand
what is fragile X? what is the repeat? what state can be detected and what can result from the mutation? how is it analyzed?
most common genetic cause of intellectual disability, caused by a mutation in the FMR1 gene on the C chromosome which leads to trinucleotide repeat expansions
repeat is a CGG triplet-repeat and is large in full fragile X that are microscopically visible
normal is 5-40 repeats, carrier is 56-200 repeats, and full mutation is 200-2000+ repeats
premutation state can be detected
expansion can result in methylation of FMR-1 and gene silencing
length of expansion analyzed through PCR-capillary electrophoresis, touchdown CPR, or southern blotting if the mosaicism must be detected
tends to use touchdown PCR and if the mosaicism still cannot be detected, use southern blotting with radiation
what is anticipation?
repeat expands every subsequent generation and the symptoms are worse and manifest earlier
what is touchdown PCR? what are the steps for touchdown PCR?
touchdown PCR utilizes a high annealing temp and slowly drops it down for more specific binding because if you go to the right annealing temp right away, there may be a lot of nonspecific binding sequences
uses high annealing temperature first (95) near the melting temperature
95 is trying to find the annealing temp where 50% of the target is bound by primers
drop the temp by a couple degrees
sweet spot = 40-50 where there is nothing but specific priming
keep dropping a couple more degrees where more of the primers are used for specific priming due to them being finite
when it is at a cooler temp, there will not be enough primers for any non-specific binding
how was fragile X detected by touchdown PCR and southern blot?
both methods use radiation
in PCR, it is less clear, normal will have smaller repeats and will be further down the gel and
bands between normal and premutation are heterozygous meaning they may have a normal child
premutations are further up the gel with 50-90 repeats
in southern blot, clearer b ut needs more radiation
full mutations have thicker bands and will be closer to the well due to the increased repeats
required to detect full mutations due to their large size

how did the trinucleotide repeat in huntingtons show a parental transmission bias in the pedigree?
father is affected and affects three of his children
daughter 2 is heterozygous and two sons have the disease
fathers symptoms presented later due to having shorter repeats presented in the bands
sons 4 and 5 had longer repeats which caused their symptoms to present earlier
son 1 will present symptoms later and will get worse as he ages
ALL SHOWN THROUGH THE BANDS
what is labelled probe PCR?
primer is labelled with radiation at the end and will flank the repeat expansion to see how big the repeat is then ran on a gel
>40 repeats = huntingtons disease
10-29 repeats = normal
what requires PAGE?
higher resolution DNA gel required PAGE
it shows the number of repeats corresponding to the ladder
old school but still the best
how many genes are expressed on the mt that are not in nDNA?
37 genes and these genes are unique only to mitochondria
what is the study in 2015 discussed in class that still works today for mitochondrial mutations?
there is a biochemical level that has to be reached and it was thought to be up to 50% before the threshold is pushed over and the phenotype is visible
means that it falls under the recessive class because the phenotype is similar to a recessive trait
how was the detection of NARP mito point mutation detected by PCR-RFLP? how was the KSS mito deletion detected by southern blotting? what happens in S.blot if you’re heterozygous?
PCR RFLP
presence of the mutation creates a Msp1 REN site in the amplicon
will have three bands for the presence of the mutation
SOUTHERN BLOTTING
the REN Pvu2 cuts once in the circular mtDNA which will detect the deletion pattern
if you have a heterozygous population, might not see smaller fragments because they aren’t that common in the mitochondria but by s. blot, it will be more sensitive than PCR-based analysis
what is clinical DNA sequencing rapidly becoming? what did they use to do and what is expensive? what can you do with NGS?
rapidly becoming the gold standard for diagnosis of mitochondrial disorders
used to do muscle biopsies but they’re painful and muscles don’t regenerate well
the actual downstream amplification is expensive because the biopsy required multiple visits with the patient but requires a good histology procedure because you can’t go back and get more
with NGS → can see whole genome in MT
can separate mtDNA and nDNA easily
now there’s kits for mt with primers and conditions which saves time
what are the three NGS kits?
courtagen life sciences
includes sequencing of either the mtDNA or the nuclear encoded mt genes
mit-o-matic
comprehensive computational pipeline for clinical evaluation of mt variations
mito-aND-panel
combined sequencing of mtDNA and nDNA
discuss the limitations of genetic testing by molecular techniques (written response question)
Intergenic mutations in the splice sites or regulatory regions can be missed when analyzing the exon regions. Sometimes, the entire genome cannot be scanned so the intergenic mutations may be missed. Most medications are used to treat symptoms not the genotype of a patient. There is a limited amount of money that can be spent on an individuals health so it may not be worth it to spend extra money to figure out the molecular background of a specific disorder if the medication is working for that patient. There also are ethical concerns when diagnosing individuals who do not have a disease phenotype yet. Lastly, most diseases and normal traits are multicomponent systems that involve genes and environmental factors.
what is the reason why there is rare penetrance in some genes?
there are so many other things in an individuals life BESIDES DNA that can affect the disorder
could have a mutation and you might or might not develop cancer
how is NGS used for genetic disorder detection?
is the holy grail for genetic disorder detection
faster sequencing
targeted amplification and SNP detection
treatment is selected based on the specific mutation found
new methods are constantly being discovered and published
what is wiskott-aldrich syndrome and how does it relate to the genetic testing registry?
can be put into the genetic testing registry and will show a genetic test for it
it is an autoimmune issue and the gene Thrombocytopenia WAS encodes the WASP protein
deletion duplication analysis of gene by comparative genome hybridization
sequence analysis of entire coding region by sanger capillary sequence
what is direct to consumer genetic testing?
provides at home genetic testing with blood or spit (CDC)
as techniques improve and get better
more companies offer genetic testing
more prenatal diagnosis is done
there is a better understanding of traits
a hope for gene therapy