Biochemistry Exam Prep
1/101
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
102 Terms
What are the two types of evolution?
DIvergent Evolution
Convergent Evolution
What are the 3 domains of living organisms
Prokaryotes (Bacteria)
Eukaryotes (Humans)
Archea
What are the two types of proteins
Orthologs (Similar ancestor, same function)
Paralogs ( Similar ancestor, different function)
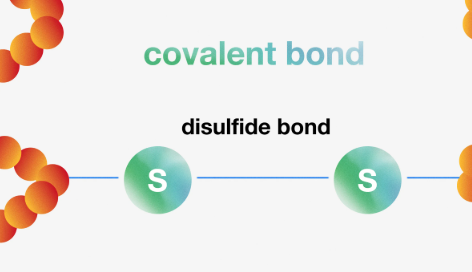
What is a disulfide bond?
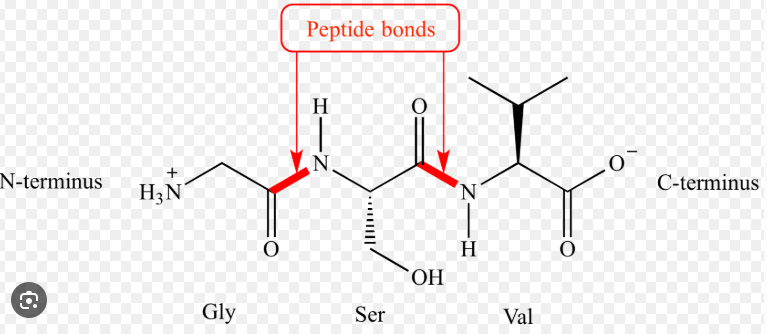
What is a peptide bond?
Explain what the hydrophobic effect is
WHen a molecule is unable to form hydrogen bonds due to its non-polar character, it rejects water.
Explain what the hydrophillic effect is
When a molecule is able to form hydrogen bonds, it likes water
What are hydrogen bonds
It is the bonding due to partial charges between N,F,O an Hydrogen H
What initiates protein folding?
The hydrophobic effect
What contains the primary structure?
The individual amino acids
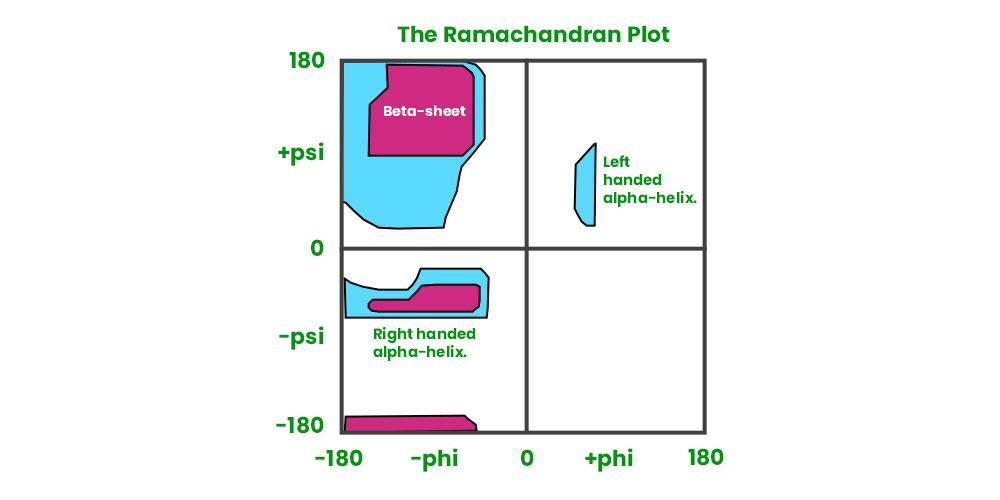
What is a ramachdran plot?
A ramachandran plot visualizes the conformations on the backbone of amino acid residues in a protein.
It has two axis:
Phi (φ) — rotation around N–Cα bond
Psi (Ψ) — rotation around Cα–C bond
What is the Alpha helix?
The helical backbone stabalized by hydrogen bonds where the side chains face outwards
What is a beta sheet
The beta sheet consists of beta strands which are the segments of the polypeptide chain.
It comes in a paralell and antiparalell version.
What are supersecondary structures?
These are extra complex structures where two helices turn around eachother
What contains the tertiary structure
Its the sum of all secondary interactions. The way protein folds into it self. Hydrophobic inside and hydroiphillic outside
Which proteins help fold proteins?
HSP’'s , Heat Shock Proteins
What is the name of the experiment that refolded ribonuclease?
The afinsen experiment, discovered HSP’'s
What contains the quartenary structure ?
The way different proteins orientate together
What are the two types of hemoglobin?
Alpha hemoglubin and Beta hemoglubin
On what basis can a protein be purified?
Size
Charge
Hydrophobicity
Binding affinity
Solubility
Density
What are the protein purification techniques?
Ion exchange chromatography
Gel filtration: makes use of inert beads to filter by size
Affinity chromatography (purifies based on affinity)
HPLC
What is His-Tag and how does it work?
His-Tag consist of 6 Histidine residues attached to the N/C terminus, it allows binding to metal ions. Which then can be eluted for purification purposes
What is protein analysis?
The act of finding out which protein you are dealing with.
What are all the different techniques for protein analysis?
SDS-PAGE
Gel-electrophoresis
Iso-electric focussing
Zonal ultra centrifusion
X-Ray crystallography
Mass Spec (Maldi-TOF)
Protein sequencing (Edman degradation)
Blotting( western, northern, southern)
Describe SDS-PAGE and how it works?
SDS PAGE stands for Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis and it seperates proteins based on thei molecular weights.
It works via the following steps:
Denaturation by SDS
Breaking the disulfide bonds making the protein into linear fragments and installing a uniform negative charge.Gel preparation
Electrophoresis
By applying a charge trhougout the gel, smaller proteins migrate faster then larger ones, creating a separation.Visualization
Proteins are stained allowing for easy visualization
How does gel electrophoresis work?
It works by applying a charge on a gel matrix and letting proteins migrate through the gel.
Explain how iso-electric focussing works
This is a technique that separates proteins based on their iso electric point. Which is the pH at which the moleucle carries no net charge.
Explain what zonal ultracentrifusion is
Centrifusion of a sample that seperates them based on density
Explain X-ray crystallography and who developed it
X-ray crystallography is a technique where a protein sample is placed on a crystal at which x-ray radiation is beamed at. This creates a pattern that represents electron density. This pattern can be turned into a map. Using fourrier transform we can reverse engineer this map to find out what protein it was.
Famous aplications which won nobel prizes are;
- DNA
- hemoglubin/myoglobun
Developed by father son bragg
Explain how MALDI-TOF works
Maldi tof stands for Matrix Assisted Laser Desorption Ionization - Time Of Flight. It identifies proteins based of their time of flight, Fragmented by a Laser in a matrix,
Explain how the edman degradation works
This is a method to analyze the sequence of a protein.
It starts at the N terminus, where a section is labelled By PTH and then cleaved. This can be repeated to create different sequences which can then be analysed.
What are the differences between wester,northern and southern blotting?
Western blotting uses antibodies to recognize proteins, making it possible to analyse and quantify protein expression.
Northern Blotting is used to identify RNA gene expressions
Southern blotting is used to identify DNA
Explain how sanger sequencing works
Sanger sequencing is a method to detect specific Nucleic Base sequences. It does this by a PCR (polymerase chain reaction) and gel electrophoresis. Each sample is stained with a fluorescent dye, a laser reads the fluorescence output and can identify which bases it comes from.
Every nucleic base shows a different light.
What are antibodies?
Invasive compounds that bind via disulfide bonds to proteins.
What is ELISA
Enzyme Linked Immuno Sorbent Assay.
It is the method of using antibodies and enzymes to identify amount of Antibodies. THe enzyme converts the substrate into a coloured product.
What are GFP’'s
Green FLuorescent proteins
WHere in the ramachandran plot can you find the right/left handed alpha helix and the beta sheet
What are the major classes of enzymes?
Oxidoreductases ( Catalyze oxidation-reduction reactions)
Transferases (Transfer a functional group from one molecule to another)
Hydrolases (catalyse hydrolysis reactions)
Lyases (catalyse breaking of chemical bonds)
Isomerases ( catalyzes the rearrangement of molecules)
Ligases (THe gluing of two molecules by atp_
What is a cofactor
the additional molecule/atom/ion required for an enzyme to function
What are the possible cofactors
metallic ions (mg2+)
Organic cofactors
What are inhibitors?
Molecules that block enzyme substrate binding
What is competitive inhibition?
It competes with the substrate for the active site on the enzyme
What is non-competitive inhibition?
This is where the inhibitor binds to a different site then the active siteW
What is suicide inhibition?
Mechanism based inhibitors that mimic a substrate but yield a inacitve product
What does the michaelis menten equation describe?
It describes how the rate of an enzyme catalyzed reaction is dependent on the substrate conncentration
What is the michaelis menten equation
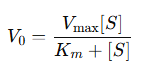
What do Km V0 Vmax represent?
Km = the substrate concentration at which the reaction rate is halved
V0 =The initial product velocity
Vmax = The maximum reaction velocity
When given a table of V0 and [S] values, you are asked to use a lineweaver burke plot to find the meachlis menten constants Km Vmax. Explain the steps needed to be taken.
Since the lineweaver burkeplot is the recipricol of the michaelis menten equation, calculate the new table with the reciprocal values.
The lineweaver burke plot is in the form y=ax+b, where a(the slope ) represents Km/Vmax.
The y intercept = 1/Vmax and the x-intercept = -1/KmFind these values and draw the lineweaver burke plot, make sure to indicate the axis and their meanings
How do the kinetics change when a inhibtor is introduced (Lineweaver burke plot)
When a competitive inhibitor is introduced the Km increases
For a noncompetitive inhibitor the Vmax decreases because this inhibitor deactivates an enzyme
What is the function of chymotrypsin
chymotrypsin is a enzyme that cleaves a disulfide bond after basic amino acids. It helps digesting protein in digestiation.
Chymotrypsin is a serine protease because serine is the most important residue.
What is a catalytic triad?
This is a group of three key amino acids found in the active site of an enzyme.
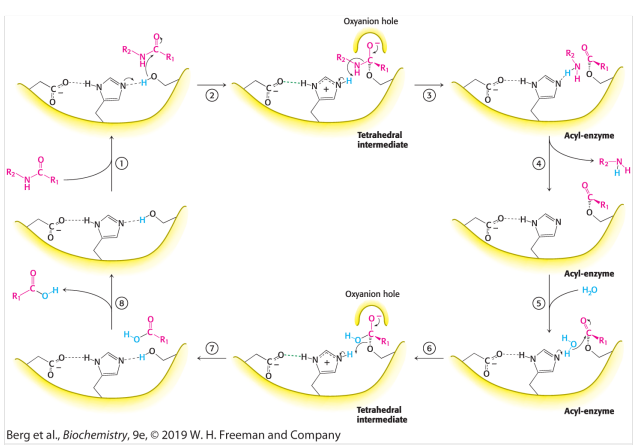
Explain the mechanism of the peptide hydrolysis by chymotrypsin
Step 1: Substrate Binding
The Enzyme-Substrate complex is formed
Step 2: Alkoxide formation
The His-Asp-Ser triad engage in hydrogen bonds making the -OH group from serine into a -O- group. This alkoxide ion acts as a nucleophile.
Step 3: Nucleophillic attack on the carbonyl carbon
The serine alkoxide attack the carbonyl peptide creating a unstable tetra hedral intermediat, This intermediate is stabalized by the Oxy-Anion Hole.
Step 4: Tetrahedral collapse
This Tetrahedral collapses forming an acyl on the serine and a amine component on the histidine. The Amine component leaves marking the End of the first part (Enzyme Acylation).
Step 5: Water Binding and Nucleophillic attack
Water binds to His and the resulting OH attacks the acyl-enzyme carbon. Forming a carboxylic acid tetrahedral stabalized by an Oxy-Anion Hole
Step 6: Tetrahedral collapse and De-Acylation
This carboxylic acid tetrahedral collapses and leaves
What other proteases are there?
Cysteine (-SH nucleophile)
Aspartyl (-H2O nucleophile)
Metallo
At which residues does Chymotrypsin cleave?
At aromatic residues like: Phenylalanine, Tyrosine, TryptoPhan
At which residues does CNBr (Cyano Bromide) cleave?
Cleaves after methionine
At which residues does trypsin cleave?
After basic residues like: Lysine and arginine
At which residues does Carboxypeptidase cleave?
It cleaves from the C-terminus
Explain how cation exchange works
Cation exchange works by seperating molecules based on their net charge, this is done by using a negatively charged resin. Negative/ neutral fragments have no affinity for this resin and elute freely. Positevly charged fragments stay behind and this separated.
Explain how monoclonal vs polyclonal antibodies can be created
monoclonal antibodies can be created by injecting an organism with the target virus. This organism then creates the B-cells that fight against that target virus. These B-cells are then collected and merged with cancer cells. These fused cells then multiple indefintely and are collected. These fused cells are then screen to find the best ones that produce the desired antibody.
What does Kcat mean?
The substrate turnover number at max velocity
What is the function of DNA polymerases?
It is an enzyme that catalyses the formation of new nucleotide chains and need a primer with a free 3’OH.
What are the functions of metal ions in DNA synthesis?
To stabalize the template
And to allow the activation of the 3’ hydroxyl group of the primer
In which direction does DNA synthesis occur?
5’ → 3’
What is the replication fork?
The site where DNA synthesis starts
What are the two fragments that get formed during DNA synthesis?
Leading strand ( nucleotides are continously added)
The lagging strand-okazaki fragments-
What is the function of DNA ligase
It glues the okazaki fragments together
What is the function of DNA helicase?
It creates the replication fork, unzipping the DNA
Explain what the term supercoiling means
Supercoiling is when DNA helices are torsionally folded into them selves creating strain on the helix. Supercoiling can be fixed by topoisomerases.
Exists in a negative and postive manner.
List all the different DNA polymerases and their functions
DNA Polymerase 1
Removes RNA primers and replaces them with DNA
DNA Polymerase 2
Repairs DNADNA Polymerase 3
Replicates DNA
List all the different Topoisomerases and their functions
Topoisomerase 1
Catalyzes the relaxation of the supercoiled dna, which is a thermodynamically favourable process due to the strain present in supercoiled DNATopoisomerase 2
Utilizes ATP to relieve supercoiling
Explain the entire process of DNA replication
DNA replication begins at the origin of replication, where a Helicase unwinds the DNA
DNA Primase then synthesise primers which DNA Polymerase 3 recognizes and starts adding nucleotides to in the 5’' → 3’' direction.
A Leading and lagging strand are then created, gaps in the lagging strand are glued together by DNA ligase
DNA Polymerase 1 replaces the primer with DNA
What is the function of RNA polymerases?
RNA polymerases catalyze the Transcription process where DNA gets turned into RNA
Where does RNA synthesis (transcription) start?
At the transcription bubble, a locally unwound piece of DNA
What is the coding strand?
The strand of DNA that has the same sequence RNA only with Uracil instead of Thymine
What are the three stages of Transcription and what happens at each stage?
Initiation
RNA polymerase binds to the promotor region and forms the trancription bubbleElongation
RNA polymerase adds nucleotides to the 3’ end creating a DNA-RNA hybridTermination
What is the difference between pre-mRNA and mRNA
pre-mRNA contains useless junk info called introns and mRNA contains only the neccesary nucleotides for the target protein
Explain what splicing is
Splicing is when the introns present on pre-mRNA get ‘‘spliced’’ or removed leaving only the exons present (mRNA).
This happens through a process called the splice mechanism
Explain in detail what the splice mechanism is
The splice mechanism is a sequencial transesterification mechanism.
A large protein complex called the spliceosome controls the process of splicing. This Spliceosome is made up of small nuclear ribonuclearproteins ( snrnp’s). ( U1,U2,U4,U5,U6)
U2 binds to the Adenosine branch site on the pre-mRNA and U1 binds to the 5’ splice site on the intron.
U4,U6 and U5 also bind to the intron creating a hairpin like structure called the lariat intermediate
The 2’OH of the A branch point attacks the 5’ splice site and it gets cleaved. U1 and U4 are released.
The newly freed 3’OH side of the exon attacks the other side of the intron cleaving that too.
The lariat is freed and the two exons join together.

What is translation ?
It is where mRNA gets turned into amino acid sequences ( protein)
What does protein synthesis require?
The recognition of three base pairs called a codon on messenger RNA which code for a specific amino acid.
What is tRNA?
Transfer RNA which holds one amino acid that recognizes a codon.
Transfer RNA serves as the adapter molecule that binds to a specific codon and brings with it an amino acid for incorporation into the polypeptide chain.
Which enzyme catalyzes the amino acid chain formation?
aminoacyl-tRNA-synthesase
Where does this Protein synthesis take place?
At the ribosome, jsut outside of the nucleus
List all the sites on a ribosome and their functions
The A site
The entry point of incoming tRNAThe P site
Holds the tRNA with the growing peptide chainThe E site (exit site)
Where the newly formed peptide chain leaves the ribosome
What are elongation factors?
Proteins that assist the ribosome in reading the codons and building the peptide chain.
What is EF-Tu?
Elongation Factor Thermo Unstable
It delivers tRNA to the A site of the ribosome
What are EF-Ts?
Elongation Factor Thermo Stable.
It recycles EF-Tu so it can participate in another round of protein synthesis
What is EF-G?
Elongation Factor G.
It uses energy to move mRNA through the ribosome
What are the analogues of EF-Tu,EF-Ts and EF-G for eukaryotes?
eEF1A,eEF1B,eEF2.
What is the endoplasmatic reticulum (ER)
Two kinds:
Rough ER
the ‘‘car park’' of ribosomesSmooth ER
Produces Lipids
What is the Lumen?
A space enclosed by the ER where proteins get folded and checked before secretion.
What is the rho protein
a transcription termination factor
What is poly adenylation?
is the process in eukarytoes where a poly(A) tail is added to pre-mRNA
What is a poly(A) tail?
THis is a sequence of adenine residues thats put on mRNA in order to protect it from exonucleases.
What is the 5’ cap and its function
It the addition of a guanine cap to the 5’ end of RNA, this cap also protects the mRNA from exonucleases
At pH = 7 which amino acids get -/+ charge
Aspartic Acid (-)
Glutamic Acid (-)
Lysine (+)
Arginine (+)
Histidine (+)
explain the process of topoisomerase 1 and 2
Topoisomerase 1 cuts a single strand dna , uncoils it need atp, and glues it. Topoisomerase 2 cuts both strands from which it uncoils itself due to strain.
Adenine
Guanine
Cytosine