Cell final review ALL
1/473
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
474 Terms
Define chromatin
A single package of DNA (half of a chromosome) and protein
Chroma (color) PLUS protein = chromatin
define heterochromatin
Chromatin that is highly condensed and contains silenced genes or very few genes
different types depending of location
Define euchromatin
Less condensed chromatin that can be packed into heterochromatin
genes close to heterochromatin (even if its suppose to be euchromatin) are affected because condensation spreads
Define polytene chromosome
Chromosomes that aid in understanding chromatin organization
histone variants and non-histone proteins makes for differential regulation of gene expression and varying roles in a cell
differences in condensation levels within a chromosome
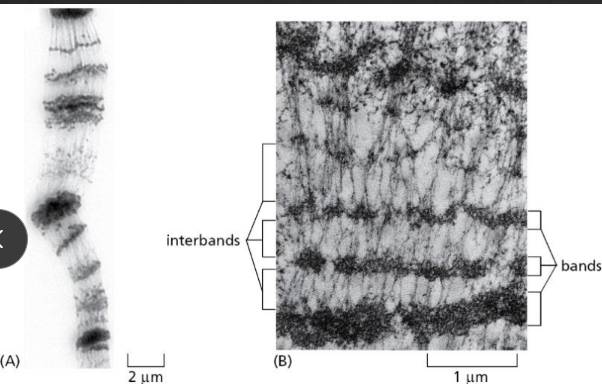
Define condensins
Large proteins that act like combs that DNA can twist around
located at the base of loops near scaffold
when gene is going to be transcribed, the area decondenses, unwinds, and transcibes
Define nucleosomes
The basic unit of chromatin structure
Made up of packed histones
each nucleosome has ~200 bp of DNA wrapped around it & contains 2 of each four histone subunits.
nucleosome is the actual circle with the DNA (histone + DNA)
Define histones
Small proteins that bind DNA via 142 hydrogen bonds
contain a lot of arginine and lysine (positively charged)
binds to negative DNA
Make up the first level of organization and are packed into nucleosomes.
Has 4 subunits and together make an octamer
Define ploidy
the number of sets of chromosomes in a cell, or in the cells of an organism.
Define conserved synteny
Parts of mammal’s genome that is contains the exact same genes in the exact same order (in humans found in other mammals)
Define fractal globule
Chromosome with a physical structure that allows it to change shape (unfold/fold & densely/loosely packed)
can fold and unfold as necessary (solenoid model)
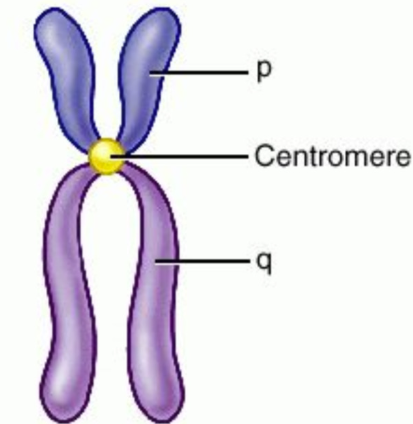
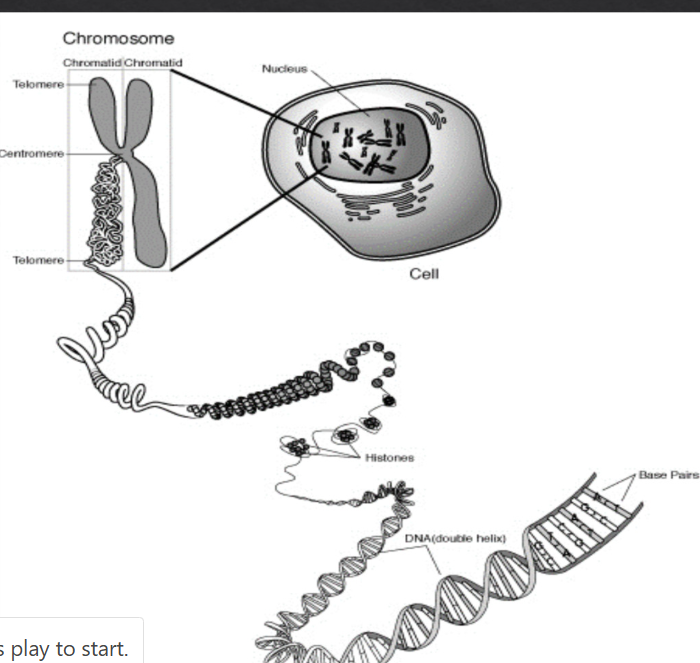
Label the 3 parts of the chromosome
Centromere = allows one copy of each duplicate to be pulled into each daughter cell
p arm = short arm
q arm = long arm
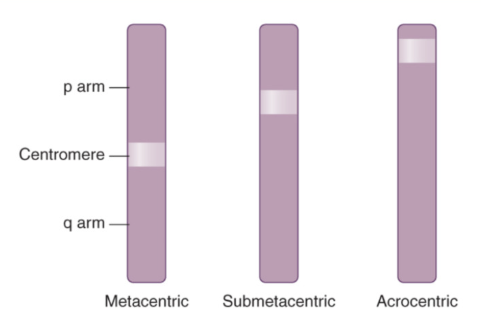
Name each type of chromosome based on the location of the centromere
Metacentric = centromere found straight in the middle of the chromatin
Submetacentric = centromere found slightly off center (different arm lengths)
Acrocentric = centromere found on the edge of the chromatin (very small p and large q)
ex: chromosomes 13, 14, 15, 21, & 22
What is important about a centromere
Centromere = allows one copy of each duplicate to be pulled into each daughter cell
Kinetochore (little protein) at the centromere forms and attaches both centromeres to the mitotic spindles
Describe transposons & its percentage
~40%
DNA that was inserted over long periods of time
usually are unused, but sometimes can insert themselves in new locations that cause a mutation or new function
“mobile genetic elements”
parasitic sequences that can disrupt function or alter gene regulation “jumping genes”
not always bad
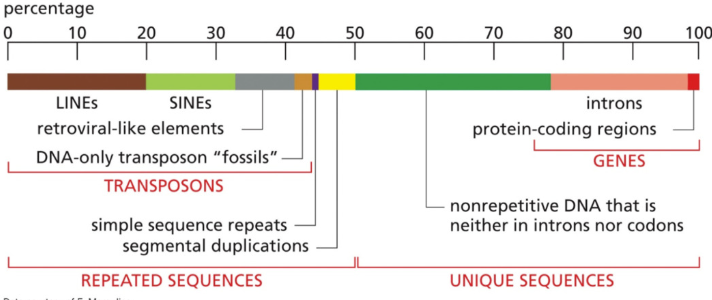
Describe introns and their percentage
~20%
Don’t code for proteins
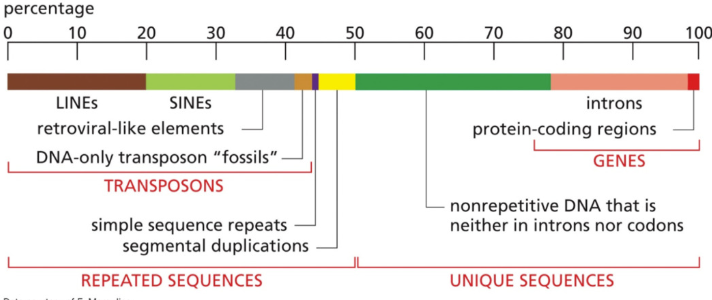
Describe exons and their percentage
~1-2%
code for proteins
Describe genes and their percentage in the genome
~20%
Code for something, have a process in the protein function/development
What are the 3 major components necessary for chromosomal function (list)
replication origins = where DNA dupication begins
there are many and all simultaneously begin replication
Centromere = allows one copy of each duplicate to be pulled into each daughter cell
Kinetochore (little protein) at the centromere forms and attaches both centromeres to the mitotic spindles
Telomeres = Repeated sequences forming the ends of a chromosome
protect it from being eaten by DNA repair genes and replication
What are the cell cycle phases & chromosomal state in each
Interphase = interphase chromosomes are replicated and are very loose
DNA easy to access
1 chromatid only
Mitotic phases (M phase) = mitotic chromosomes are now highly condensed so they can be separated/distributed into 2 daughter nuclei
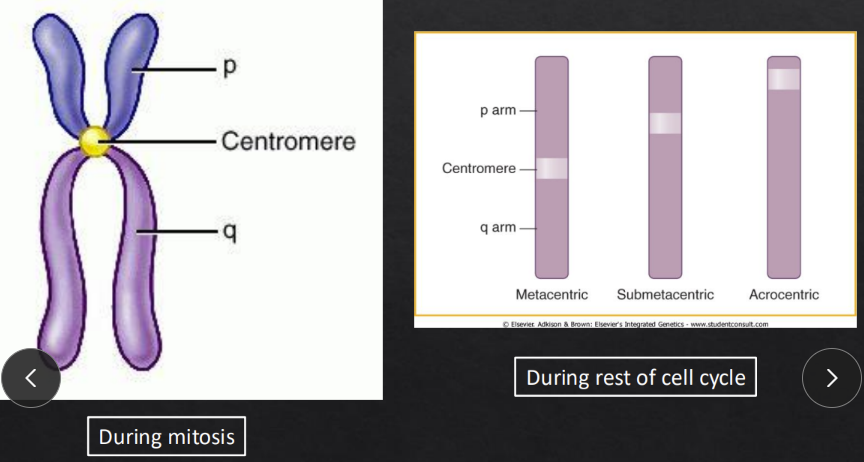
How do chromosomes change states (compaction level) between cell cycle states and proteins involved
Condenses are large proteins that further condense the fiber loops
when a gene needs to be expressed, it unwinds
histones are the octamer proteins that begin the condensing process
What is a nucleosome? It’s function, proteins involved in packaging DNA, and structure
Nucleosomes = the basic unit of chromatin structure
DNA wound around a histone core (THE BEAD)
200bp of DNA wrapped around it
contains 2 of each histone subunit (histone octomer)
held together via 142 hydrogen bonds
makes DNA 1/3 of it’s original length
What are the type nucleosome packaging models?
Space-filling model
nucleosome made up 4 histone types (proteins)
four histone types bind to DNA with hydrogen bonds
histones have a lot of arginine and lysine (positive charge) that is attracted to DNA’s negative charge
Zig zag model
Nucleosomes structured into a chromatin fiber
Solenoid model
chromosome folds and unfolds into densely or loosely packed chromatin making a fractal globule
Has a physical structure that allows it to change shape
State the role of histones and chromatin forms in cells, including structure and function
Histones = first level of organization
packs into nucleosomes
Chromatin
Fiber with a 30nm diameter
Can form chromatin loops that can range between 50k-200k
chromatin loops
Can decondence and condence with condensin proteins (when unwound, the region can be transcribed/active transcription)
Define and explain chromatin/chromosome territories within a cell nucleus
Interphase chromosomes occupy discrete territories in the cell nucleus; that is, they are not extensively intertwined.
chromatin moves around in the nucleus (territories can change depending on the level of expression required of the gene)
gene rich regions tend to move centrally in the nucleus
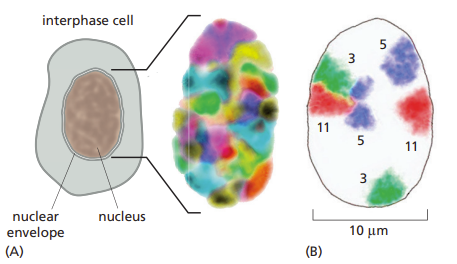
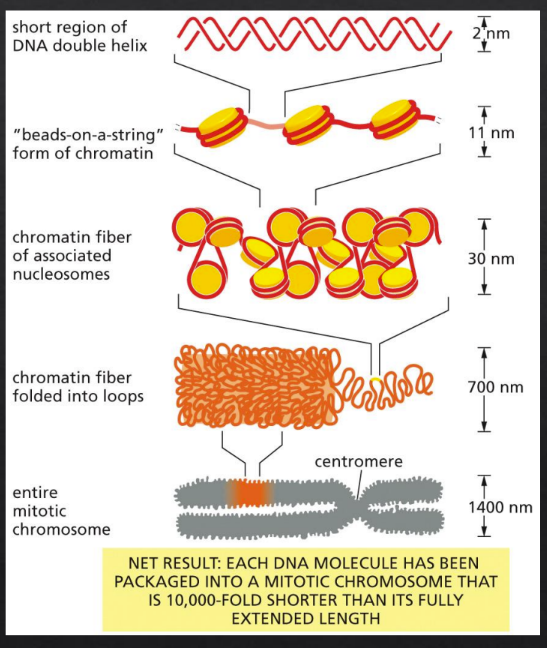
Describe the packing and organization of DNA in chromosomes (include timing during cell cycle & proteins involved)
DNA
Histone
nucleosomes
30nm fiber
loop domains
chromatin fiber
Mitotic chromosome
What is the process of DNA replication (eukaryotic)
Origin of replication
DNA replication process: (on loose DNA)
Initiator proteins (ORC) bind to origin
The origin is usually a sequence rich in ATs
Chromatin structure/transcriptional activity may define origins
DNA/protein binding at origin calls over the helicase
Helicase unwinds the DNA when it is phosphorylated
Both the ORC and helicase are phosphorylated
ORC is inactive
Helicase is active (begins unwinding)
Primase comes and makes the RNA primer
DNA polymerase alpha
DNA polymerase ε begins transcribing the leading strand & DNA polymerase δ makes the lagging strand
Nucleosome/histone reassembly (semi-conservative)
Increased mRNA histone transcription & decreased mRNA destruction
Preparing for M phase (which is highly condensed) & double amount of DNA
List the cellular mechanisms that help avoid error generation during DNA synthesis
ORC being highly regulated
Phosphorylation controlling activation/inactivation
Mismatch repair genes are signaled by “nicks” in DNA to come and fix it
Strand directed mismatch repair
Muts comes to error, determines which is the daughter/parent strand, cuts out error, DNA polymerase comes and synthesis
DNA polymerase selectivity/proofreading
Cell cycle checkpoints
DNA repair pathways
How do the phases of the cell cycle relate to DNA replication
G1 phase = ORC sets up (everything is brought)
Phosphorylate helicase/ORC to begin replication
S phase = replication only occurs during S phase (in interphase)
Everything gets replicated
G2 phase = repairs any DNA mistakes made during S phase
M phase (mitosis) = separation/daughter cell forms
Define telomeres
repeated ends that are a “counting mechanism” to avoid unwanted cell proliferation
Repeated sequence (TTAGGG) at ends
When ends get too short (can no longer keep up with chromosome duplication) the cell stops dividing and dies
In cancers, there can be issues with telomerase
Disease with dysfunctional replication
Trinucleotide (triplet) repeats = replication errors
Di/trinucleotides located in repetitive sequences in an unstable area leading to slippage and expansion during replication
Lagging strand will make a bubble -> to expanded sequence
BER can keep this from happening
Ex: Huntington’s, FXS, and myotonic dystrophy
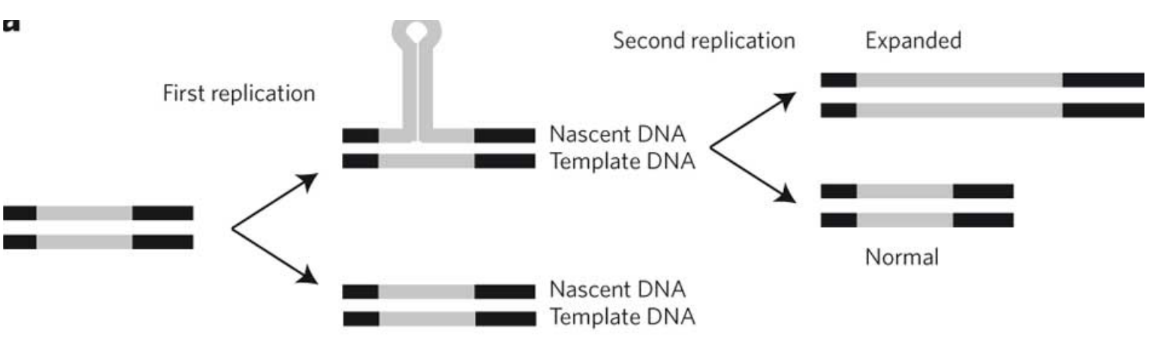
What enzyme makes/assembles telomeres
Telomerase = replicate the ends of chromosomes (reverse transcriptase)
3’ end always longer, therefore tucks into t-loop (double DNA strand)
What is one disease that is due too telomerase issues
Dyskeratosis congenita = abnormally short telomeres
Missing nails, alopecia, and abnormal skin pigmentation
Define germ cell
transmit genetic info from parent to offspring
Contain heritable information, so it WILL affect the offspring
Ex: eggs/sperm
Define somatic cell
form the body
Mutations will only affect the body and NOT the offspring
Define mutation
permanent change in DNA
Can be good or bad
Mutation rate = rate at which observable changes occur in DNA sequence
~1 nucleotide change per 1 billion nucleotides
Advantage mutation = ability to proliferate extensively and spread to unknown foreign areas of host
Survive at expense of host
Define replication fork
where helicase unwinds/opens strands and replication moves along the parental DNA double strands
Active region of replication
RNA primer required
Produces 2 asymmetric strands (leading/lagging strands)
Leading strand synthesis precedes lagging strand
ORC primarily found in AT rich sites and euchromatin regions
Define replication bubble
a region of the DNA double helix that has unwound and opened to allow for DNA replication
both of the replication fork areas (opened)
Define DNA polymerase
an enzyme that synthesizes DNA molecules using existing DNA as a template
Define the leading strand
Strand that’s continuously synthesized by DNA
Define the lagging strand
Strand that is synthesized discontinuously in fragments
The fragments are known as Okazaki fragments – leaked together with DNA polymerase
Define ligase
seals DNA strands (or DNA areas) together
Define primase
adds RNA primer
Acts as guide
Part of replication machinery
Define helicase
breaks strands apart
Breaks hydrogen bonds
Usually start at AT sites because they are “easier” to break
Part of replication machinery
Define topoisomerase
keeps DNA double strand from becoming tangled as it unwinds
Part of replication machinery
Define Okazaki fragment
short, newly synthesized segments of DNA on the lagging strand during DNA replication
Define pre-replicative complex
a protein complex that forms at the origin of replication, a specific DNA sequence, in eukaryotic cells
different from the ORC
Define telomerase
an enzyme that adds nucleotides to telomeres
added to the ends of DNA - to maintain the length of telomeres
What are the functions of DNA polymerase
Synthesize new DNA strands, ensuring accurate replication and the maintenance of genetic information
SERVES AS A PROOFREADER & ERROR CORRECTOR
Finishes Okazaki fragments & leading strands
provides exonucleolytic activity to remove mismatched bases
drives DNA repair with the energy of phosphodiester bond formation
Family A
Pol y = replicates and repairs mitochondrial DNA
Family B
Pol α = starts RNA primer synthesis (primase)
Pol δ = makes lagging strand
Pol ε = makes leading strand
Family X
Pol β = DNA repair
Pol λ & µ = NHEJ repair
Family Y
Pol n, l, & k = Trans lesion synthesis (push through errors)
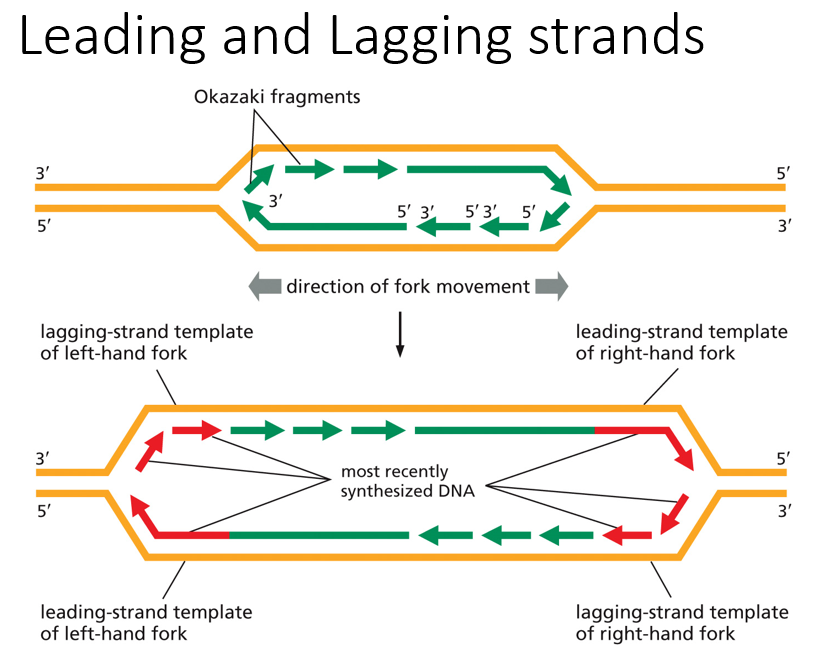
Identify the leading/lagging strands
In what direction is DNA synthesized
DNA is synthesized 5’ to 3’
What is an aneuploidy
loss/gain chr.
ex: Down syndrome, Turner’s, & Klinefelter’s
What is a isochrome formation
two of the same arm – q or p
one arm is lost and the other is copied (replaces the missing arm)
Ex: Angelman disease
What is Cri-du chat disease
Deletion on short arm of chromosome 5 (arm p)
Deletion of 5p
Profound retardation and cat like speaking (issues with larynx)
What is down syndrome
Autosomal aneuploidy (trisomy)
3 chromosome 21
* Isochrome of chromosome 21 (2 q arms) can have similar phenotype to DS
What is Klinefelter’s
WSex chromosome aneuploidy (trisomy)
XXY
Mixture of female/male characteristics (usually sterile)
What is Turner’s syndrome
Sex chromosome aneuploidy (monosomy)
XO
Short stature, underdeveloped sexual characteristics (usually sterile)
What is CML (Chronic Myelogenous Leukemia)
Translocation between chromosome 9 & 22
The ends of both chromosomes q arms are translocated
Bone marrow disease characterized by increased WBC line proliferation
What is Angelman disease
Isochrome of chromosome 15 (two maternal q arms)
The paternal chromosome is normal; the maternal chromosome is the one with the isochrome characteristics (2 chromosome 15 q arms)
Define antiparallel
the two strands of the double helix run in opposite directions
Define base
A nitrogen-containing rings
Purine or pyrimidine
Chemically/physically distinguishable
CuT = pyrimidine
Au:Gold = purines
Define base pair
Purines CANNOT bind to other purines because of chemical structure
Keto group of one base binds to amino group of another
Define complementarity
one strand complements the other
Necessary because of base pairing requirements (hydrogen bond formation)
Define double helix
a pair of antiparallel helices intertwined about a common axis, especially that in the structure of the DNA molecule.
Define template
A pair of parallel helices intertwined about a common axis, especially that in the structure of the DNA molecule.
A DNA mold that codes for other DNA or proteins
Define intron
part of a sequence that is not used to make a protein structure
“unused” - but does have some unknown function
Define exon
part of a sequence that is used/transcribed into mRNA or eventually proteins
Spliced together to form a functional gene
What type of bond exists between bases/across DNA
Hydrogen bonds
contribute to double helix stability
What type of bonds are between sugar and phosphate in DNA backbone?
Covalent bonds
strong chemical bond with a sharing of electron pairs with a balance of attractive and repulsive forces
How many hydrogens bonds are between C-G and A-T
3 and 2
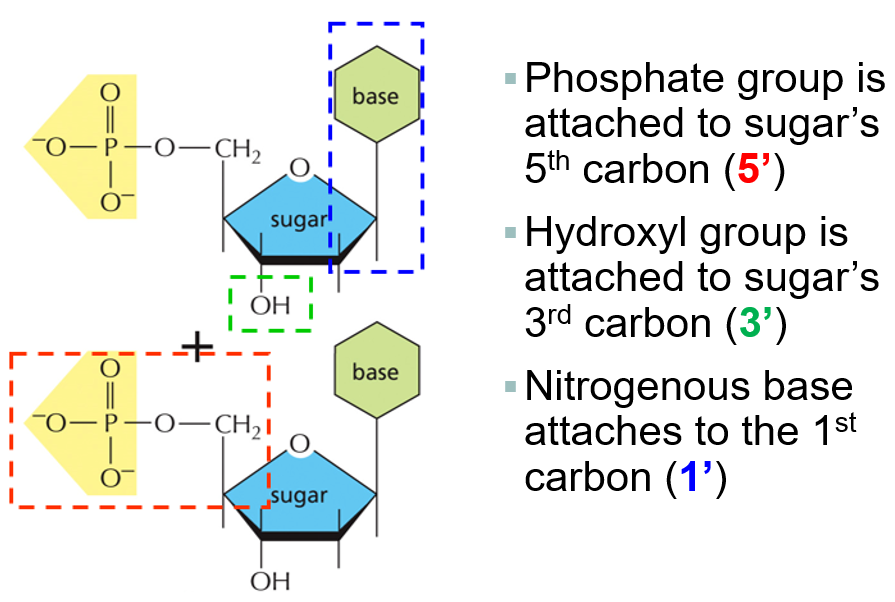
What makes up a nucleotide
nitrogenous base, five carbon sugar, and phosphate group
Sugar and phosphate form DNA back bone with covalent bonds
Where are phosphodiester bonds found
between the sugar and phosphate group in backbone
nucleotides are joined by the PO4 of one nucleotide attaching to the 3’ carbon of the next nucleotide to form a polynucleotide
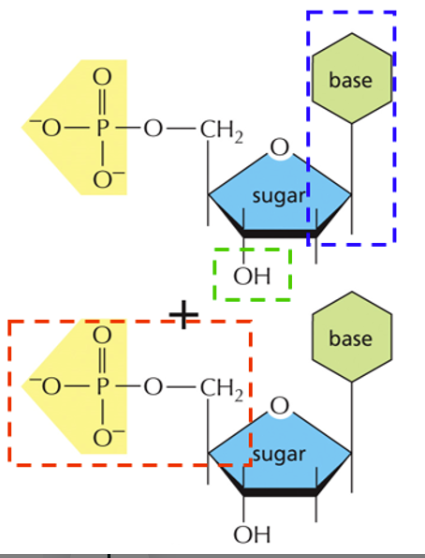
Identify the boxes and the bonds
See the image
What is the function of DNA in the overall physiology of the body
Purpose of DNA = to archive information
Not functional without it forming RNA
Is the blueprint for everything else
Protected in the nuclear membrane so it doesn’t get transported out or damaged
Only 1% of human genome is protein coding
What are universal features of ALL cells
The universal features of ALL cells:
DNA is the hereditary storage of information
Cells replicate through template polymerization (using DNA to undergo synthesis)
Use RNA as intermediate via transcription (proteins aren’t always end product)
Use proteins as enzyme catalysts to form new DNA
Use tRNA to translate RNA into protein
Proteins encoded by specific genes
Free energy (ATP) required for many cell processes)
Have plasma membrane
Can have as little as 300 genes
Define DNA
Double stranded long polymer chain composed of sequences of four monomers (hereditary information storage) double helix with bases attached via hydrogen bonds (hydrogen bonds across nucleotides/bases)
Define RNA
single stranded polymer that is flexible and can fold back on itself & can complementarily bind to other molecules/sequences
Define DNA replication
Copying (template polymerization) single strand of DNA to form another DNA strand (hydrogen bonds between bases of strands are broken)
Define templated polymerization
Using 1 copy of a DNA/RNA strand to make a new complimentary DNA/RNA strand
Define transcription
RNA synthesis. Making mRNA from a DNA strand (first step of protein synthesis)
Define translation
Protein synthesis. Making a protein from a mRNA strand
Define Codon
three nucleotides that are transcribed into an amino acid by transcribing the mRNA sequence
Define nucleotide
a sugar with phosphate group attached to a base (a single monomer in a single strand of DNA)
Define genetic redundancy
several codons can make the same amino acid (helps avoid translating point mutations)
Define a gene
a fundamental unit of inheritance, a segment of DNA or RNA that carries the code for a specific protein or RNA molecule
What are 3 differences between DNA and RNA
DNA | RNA |
|
|
What is an autocatalytic
protein with self-replicating feedback loop
Pathway makes protein and then the protein goes back and promotes more protein synthesis (or a stop)
What is a lysozyme
an enzyme that breaks apart polysaccharide chains (degrades other compounds)
Stored in the lysosome, protected by the membrane, so it doesn’t affect other processes within the cell
How can proteins direct cellular processing?
By acting as a catalysts/enzyme
Ex: Lysozymes
What are some lysosomal storage diseases
Gaucher disease, Tay-sachs, metachromatic leukodystrophy, hurler syndromes
What is Gaucher disease
Deficiency of beta-glucosidase => glycosphingolipids accumulate => enlarged liver/spleen (Gaucher cells appear in bone marrow)
RETICULOENDOTHELIA SYSTEM (RES) DISEASE (phagocytes in immune system)
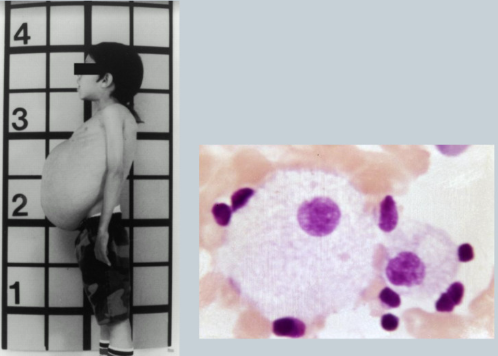
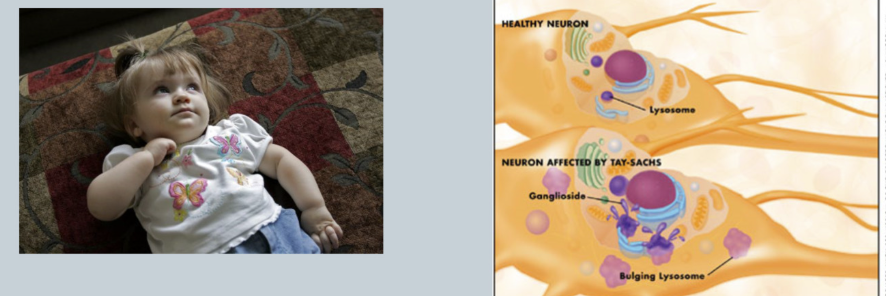
What is Tay-Sachs
Deficiency of hexosaminidase => accumulation of gangliosides in brain (break down cells) => mental retardation (hearing/vision loss)
CNS DISEASE
Common in ASHKENAZIC JEWS
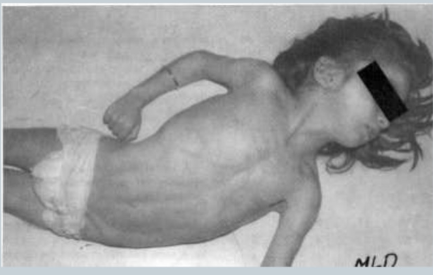
What is Metachromatic leukodystrophy
Deficiency of arylsulfatase A => buildup of glycosphingolipid in white matter of brain and cord => damages neurons => hyper flexed muscles
CNS DISEASE
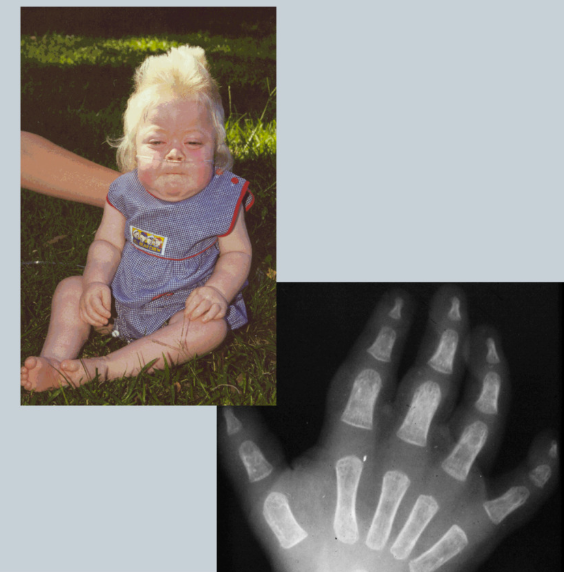
What is Hurler syndromes
Deficiency in alpha iduronidase => lack of collagen => lack of structural formation
Can’t break down Mucopolysaccharidoses because lack of lysozymes (build up)
SKELETAL & COLLAGEN DISEASE
What is the basic structures of mammalian eukaryotic cells
More elaborate structures (organelles, compartments, membranes)
Nucleolus, nuclear envelope, mitochondria, nucleus, Golgi apparatus, microtubules
Define regulatory proteins
Proteins that regulate other proteins’ production/synthesis by binding to regulatory sequences
Coding region -> mRNA -> protein -> binds to regulatory sequence -> affects protein production
Define genome
all of the genetic makeup of an organism (all of the DNA)
The entire DNA sequence of an organism
Define prokaryote
singled cell organism that can live independently and have no organelles or nucleus (free floating DNA)
Define eukaryote
complex cell that contains organelles, cytoskeleton, and has the ability to phagocytose things
Gene transcription
process of copying a segment of DNA into RNA for the purpose of gene expression.
What are key differences between prokaryotes and eukaryotes
Prokaryotes | Eukaryotes |
|
|