Active site, specificity, rate enhancement and transition state
1/20
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
21 Terms
Why are enzymes much bigger than their substrates?
Rest of the 3d structure is required in order to hold those required active site amino acid residues in the right place in the right orientation
Important in other regions og protein for regualtion - small molecuels can bind or other proteins
What is and what does chymotrypsin do?
An enzyme which digests protein
Why are the key residues that come together at the active site that are critical for catalysis' not close to each other in the primary sequence?
It's the folded 3 dimensional structure that brings them together in the correct orientation for them to bind the substrate and carry out catalysis'
What is catalysis?
The acceleration of a chemical reaction rate by a molecule that is unchanged by participating in the reaction (the process of enzyme performance) - catalyst
What takes place at the active site?
Substrate binding
Subsequent catalysis
What are active sites?
A cleft that often exclude water (but sometimes important cofactor for enzymes in the reaction)
Where do the side chains fro active site come from?
From different parts of the polypeptide chain
What are remaining amino acids in polypeptide chain which ar not needed at active site used for?
Maintain 3D conformation of active site
Provide regulatory and other binding sites
Are all enzymes specific to catalyse only one type of reaction?
Most but not all
E.g. some peptidase (break peptide bond) are also esterase's (break ester bond) e.g. chymotrypsin
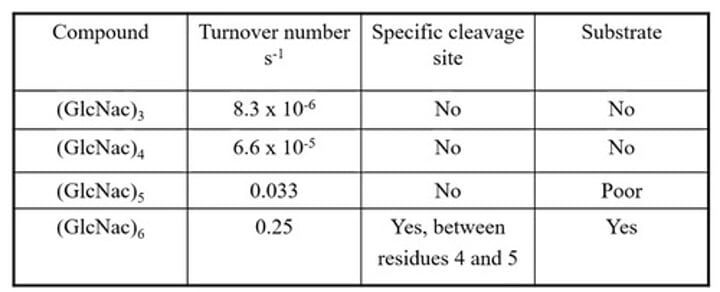
It's there an bsolute cut-off between something being a substrate or a non-substrate?
Not all enzymes are either substrate or non-substrate (no absolute cut-off between substrate and non-substrate)
Certain compounds sizes can be cut by its enzyme other sizes will not
E.h. Lysozyme activity on polymers of N-acetylcholine glucosamide (GlcNac)
2 models that explain substrate specificity?
Lock and key model
Induced fit model
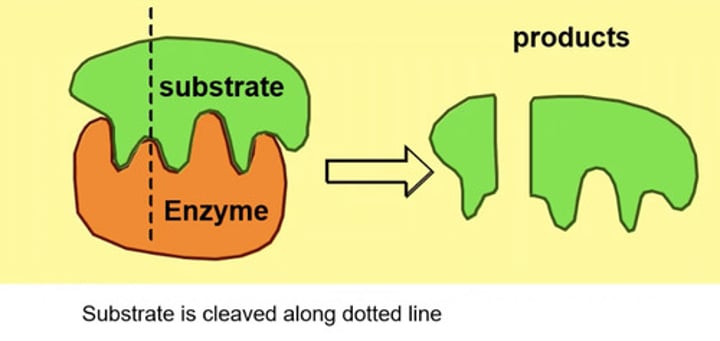
What does lock and key model state?
Substrate is specific and fifties perfectly with enzymes to form complex
Problem: molecules that have a susceptible bond should fit into active site are not substrate
What does induced fit model explain?
chemical properties of the active site of an enzyme attract the substrate, and the active site changes its shape according to the shape of the substrate - transition state
Small molecules wont induce change, so not substrate despite having susceptible bond
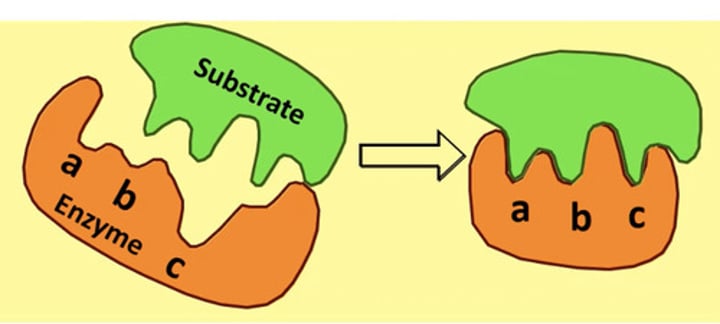
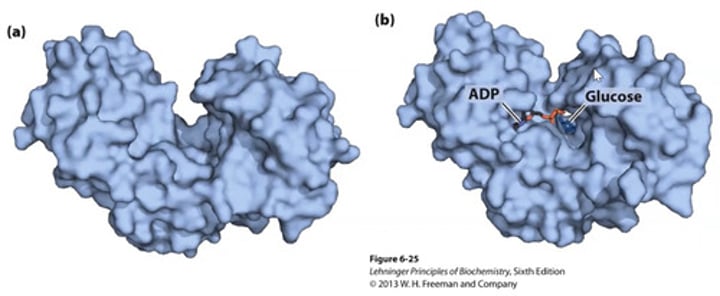
Evidence for induced fit model?
Hexokinase 'wraps around' its substrate glucose
Another reason for substrate specificity?
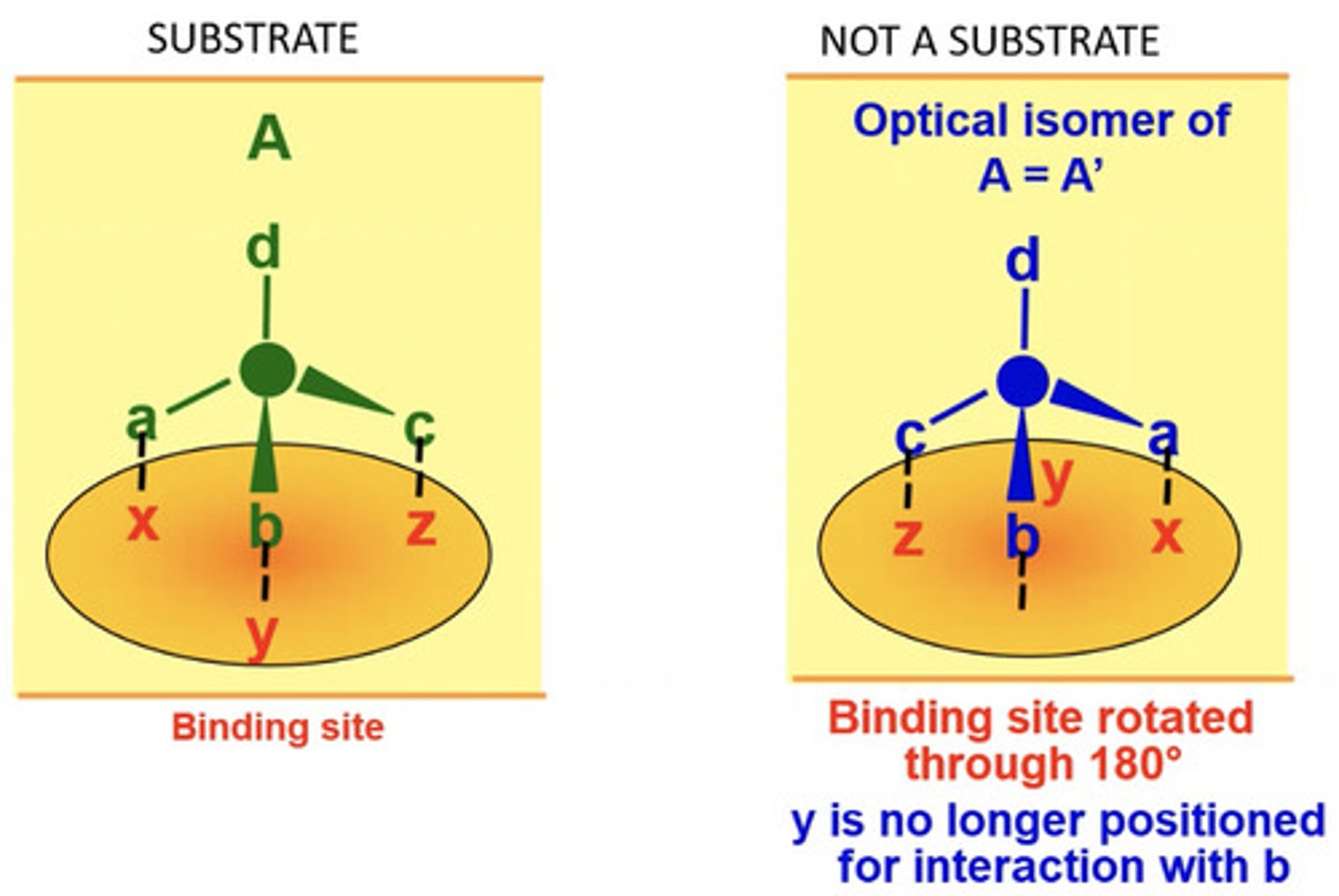
Stereospecificity
Enzymes will only bind to one of the optical isomers of a substrate - product will only be one type of optical isomer
Why is enzyme stereospecificity important in biotechnology?
Previously used chemical synthesis with rhodium catalyst - get mixture of both optical isomers - difficult to separated due to similarity in properties - important as one isomer can be helpful other one can be damaging (side effects)
Switched to enzymes - can only bind to one optical isomer - therefore produce one optical isomer - no longer need to separate them - saves company money + safer.
Why are enzymes only involved in rate enhancement?
Enzymes can only catalyse reactions which occur spontaneously in nature
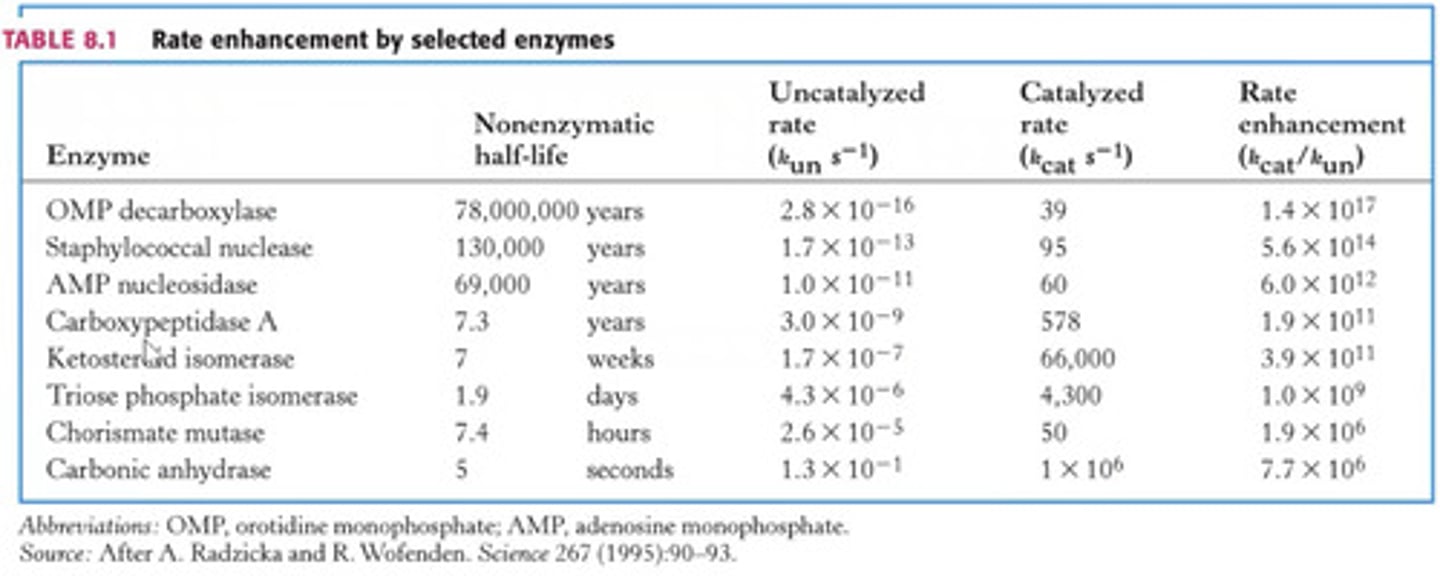
How is rate enhancement obtained?
From ratio of the catalysed to the in-catalysed rate constant kcat/kun
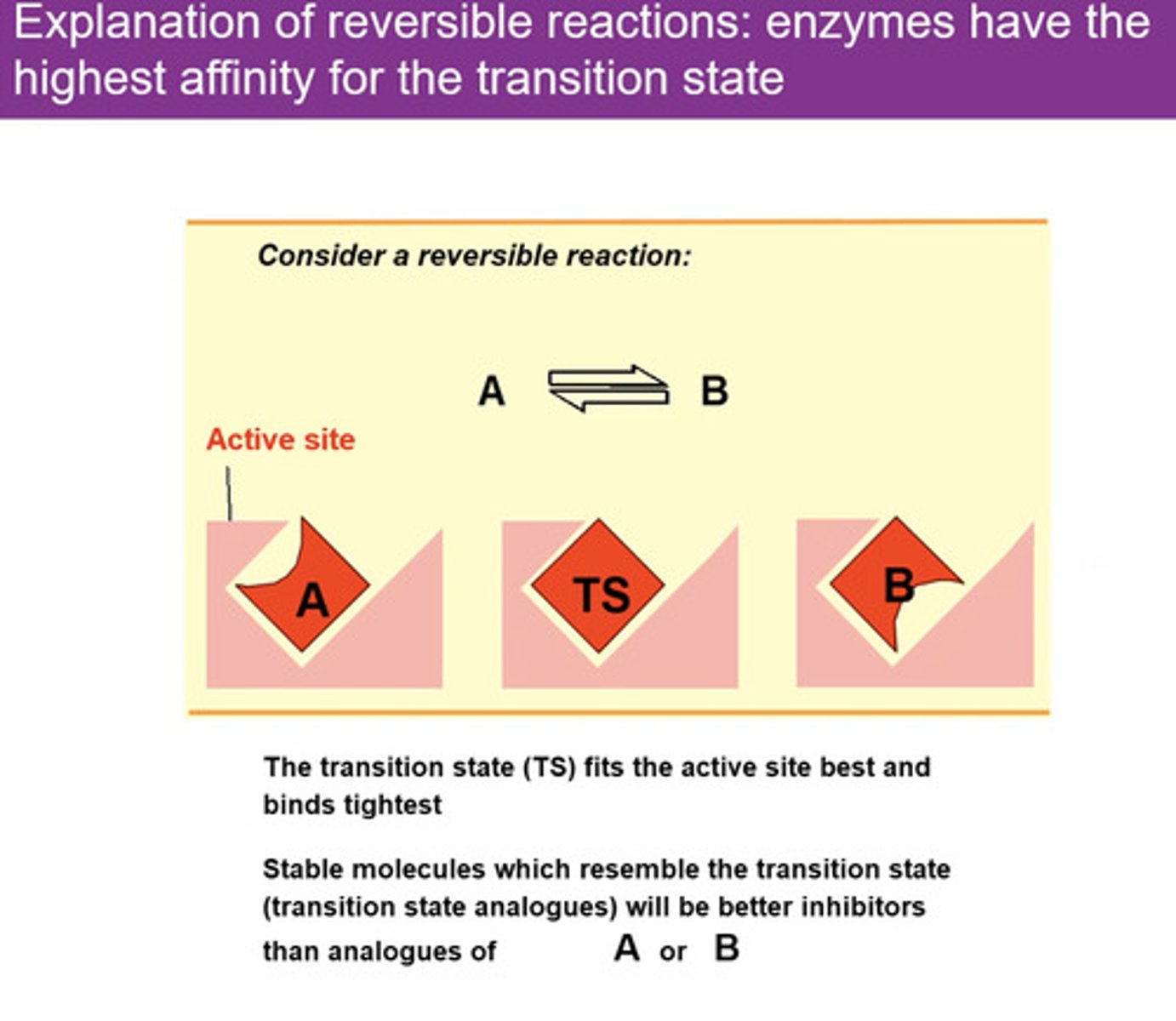
How do enzymes speed up reactions so well?
Reactions proceed via a transition state.
•The transition state is the point of highest energy on the progress of reaction curve. It is where bond breakage and formation occurs.
•The difference in energy between the substrate(s) and the transitions state is the free energy of activation
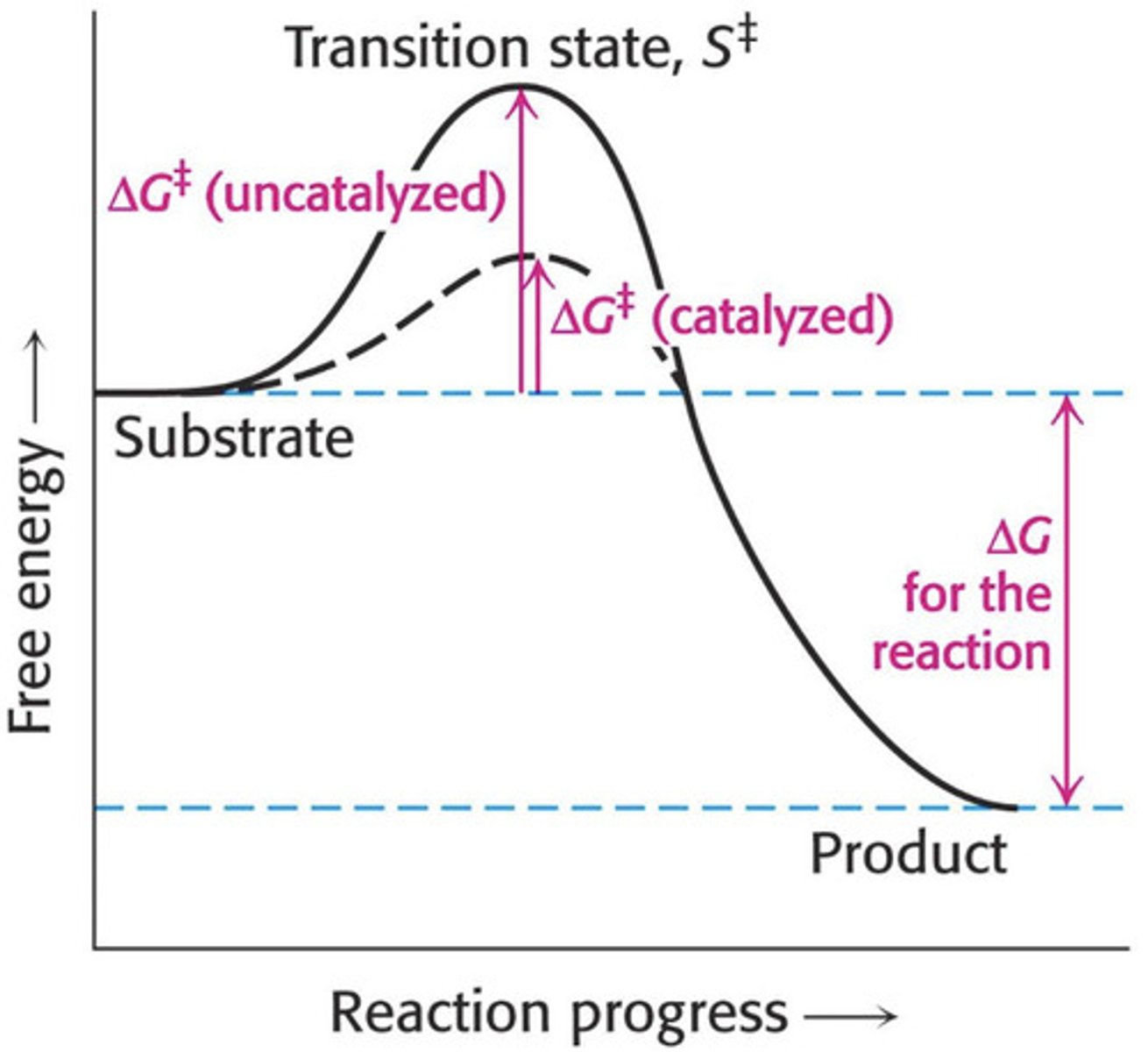
Why do enzymes speed up reactions?
Reduce free energy of activation (activation energy)
Reduce energy needed to reach transition state (i.e. stabilising)
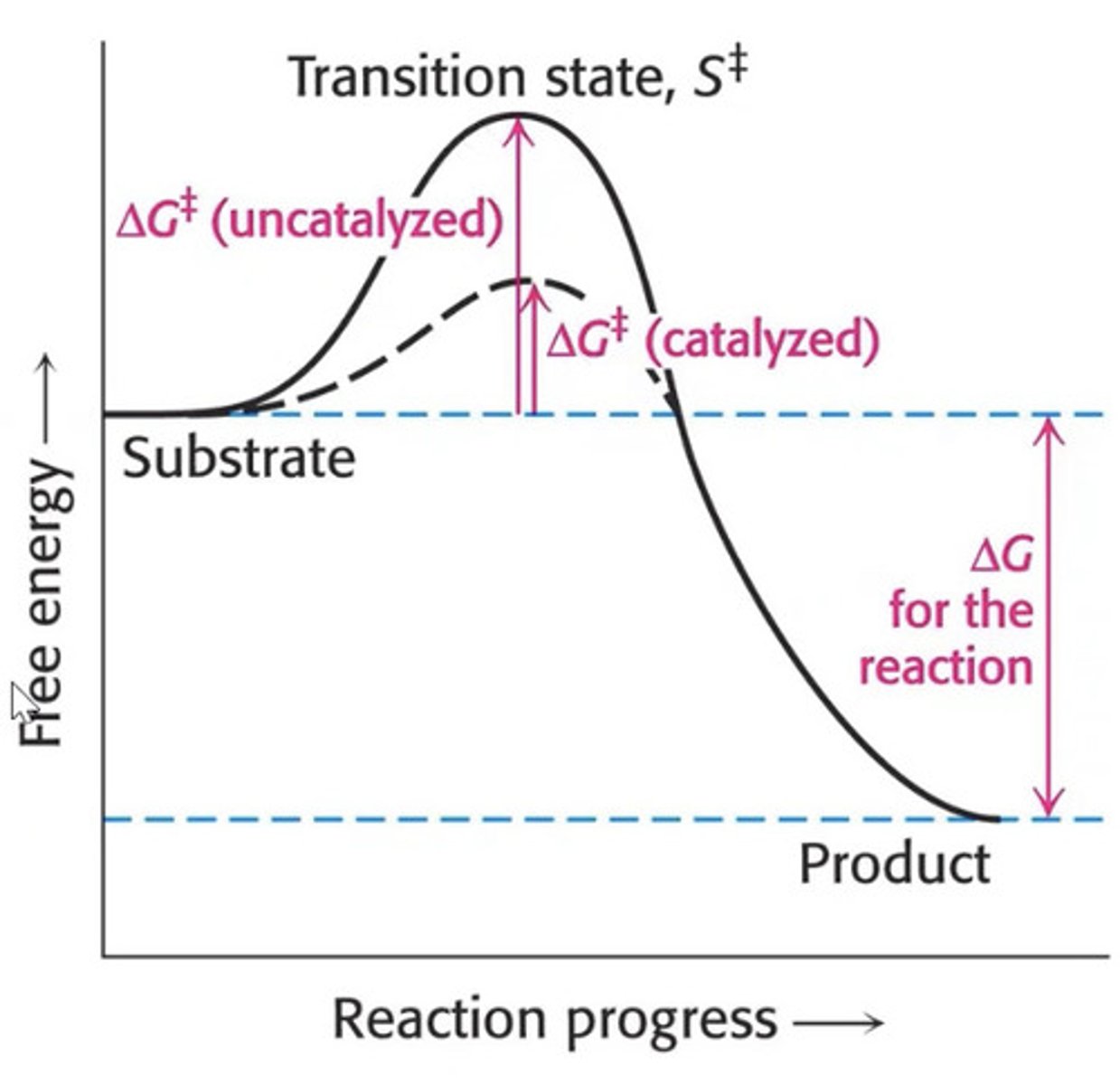
??