POPULATION GENETICS
1/50
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
51 Terms
Population
group of individuals belonging to same species
live in same geographic area
actually or potentially interbred
Population genetics
the study of allele frequencies in a population
Gene pool
all of the alleles in a reproducing population
Why measure genetic variation
determine the potential for adaptation and evolutionary change
provides evidence about the roles of various evolutionary processes
some processes increase variation, others decrease it
tells us about speciation & extinction
allows us to predict a population’s chances for long term survival
Genotypic frequency
the proportion of a specific genotype w/in a population
ranges from 0 to 1
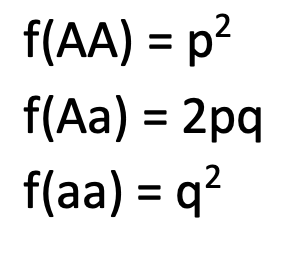
Allelic frequency
w/in a population, proportion of individuals w/ a specific allele
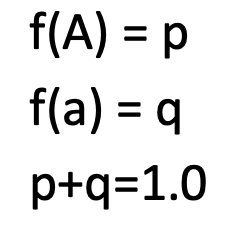
p + q
= 1… always equal to 1 in case of complete dominance
p = frequency of dominant allele
q = frequency of recessive allele
Hardy-Weinberg equation
p2+2pq+q2= 1.0
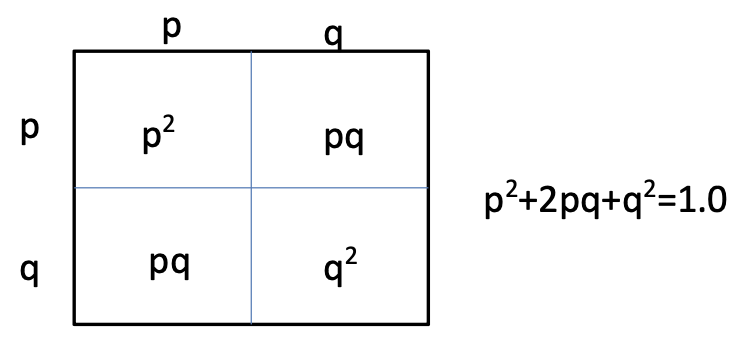
p
frequency of one allele (dominant)
q
frequency of other allele (recessive)
p2
frequency of homozygous dominant genotype
q2
frequency of homozygous recessive genotype
2pq
frequency of heterozygous genotype
Hardy-Weinberg law
allows us to calculate allelic and genotypic frequencies in the absence of evolutionary forces
gives us an idea of genetic variation in a population
assumptions about the population
infinitely large
random mating
free from mutation
no migration
no natural selection
if these conditions are met, the population is in genetic equilibrium
therefore, the frequency of alleles don’t change over time
Molecular evolution
changes in allele frequencies is due to mutation followed by natural selection or drift
most mutations are neutral
mutations leading to amino acid substitutions are usually deleterious and selected against (not usually favorable) in the environment
protein variations maintained by adaptation to certain environmental conditions
Population genetics is often used to study evolution
most populations have a lot of genetic diversity
genetic diversity allows for adaptation of the population as the environment changes
microevolution
macroevolution
Microevolution
change in allele frequencies over time within a population of a species
Macroevolution
change that results in reproductive isolation between or among populations
change leading to emergence of new species (speciation) and other taxonomic groups
Reproductive isolating mechanisms
biological barriers that prevent or reduce interbreeding and therefore exchange of alleles between populations or species
2 major types
prezygotic isolating mechanism
postzygotic isolating mechanism
Prezygotic isolating mechanism
prevents individuals from mating in the first place
mechanisms could be ecological, behavioral, seasonal, mechanical, physiological
Postzygotic isolating mechanisms
creates reproductive isolation even when members of two populations mate with each other
the resulting hybrid isn’t viable or is sterile
Natural selection
individuals with alleles that confer a reproductive advantage in the environment produce more offspring on average than others in the population
some phenotypes are more successful at survival and reproduce at higher rates
variations are heritable (passed on)
frequency of alleles that confer increased reproduction (which may or may not also be an increase in survival) increase in the population over time
ex: lactose tolerance versus intolerance
Fitness - the relative reproductive ability of a genotype
not the individual’s ability to survive, but the ability to reproduce before death
Types of selection
directional selection
stabilizing selection
disruptive selection
Directional selection
phenotypes at one end of a spectrum become selected for or against
usually as a result of changes in environment
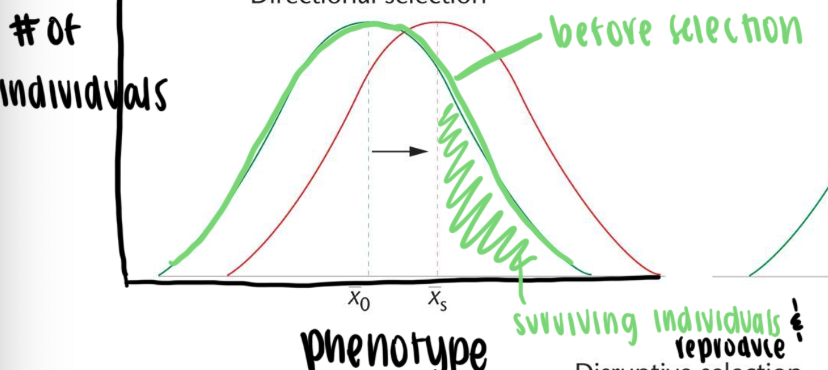
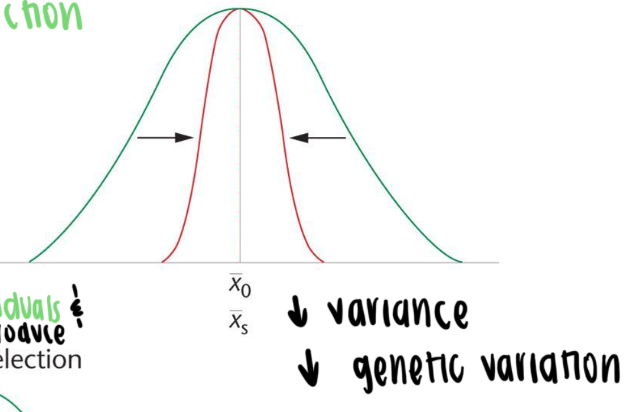
Stabilizing selection
intermediate types are favored (mean is favored)
both extreme phenotypes are selected against
reduces population variance over time but not the mean
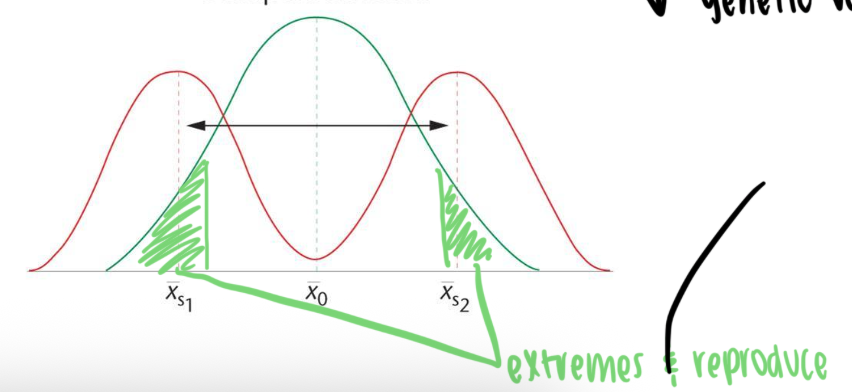
Disruptive selection
both phenotypic extremes are selected for at the expense of the mean
results in population with increasingly bimodal distribution for trait
Geographic-dependent allelic variation
allele frequencies can vary for populations separated by space or across a geographic transect
Cline
gradient for allele frequencies that change in a systematic way according to the physical attributes of an environment
Selective (nonrandom) mating
most organisms don’t mate randomly
related to natural selection
Forms of nonrandom mating
positive assortative mating
negative assortative mating
inbreeding
Positive assortative mating
similar genotypes more likely to mate than dissimilar ones
Negative assortative mating
dissimilar genotypes are more likely to mate than similar one
Inbreeding
related individuals mate
consanguineous mating
increases proportion of homozygotes in population
recessive traits are more likely to be observed
completely inbred population theoretically will consist only of homozygotes
Small effective population size
can decrease genetic variation
Effective population
individuals contributing alleles to the next generation
Bottleneck
when an effective population is drastically reduced in size and then population size rises again
selection can be natural or man-made
can result in inbreeding in a population due to low genetic variation
Migration
gene flow
genetic island
most populations aren’t completely isolated
immigration can introduce new alleles
emigration can remove alleles
alters the frequency of existing alleles
Gene flow
an organism migrates and contributes their alleles to the gene pool of a recipient population or removes alleles from the gene pool if leaving the population
Genetic island
population that breeds within itself and has little gene flow
Founder effect
when a population is established from a small number of breeding individuals
later gene pool contains only those alleles from the original population
chance can play a significant role in alleles present in founders
Genetic drift
changes in allele frequencies due to random sampling error
NOT related to natural selection
due to chance alone – no selective pressure
small populations are especially prone to drift
small effective population, bottleneck, and founder effect can contribute to genetic drift
note: these forces can also be related to natural selection
reduces genetic variation in a population
Phylogeny
genetic differences among present-day species can be used to reconstruct their evolutionary histories
differences in DNA sequence between species are proportional to evolutionary distance
Phylogenetic trees
branches represent lineages over time
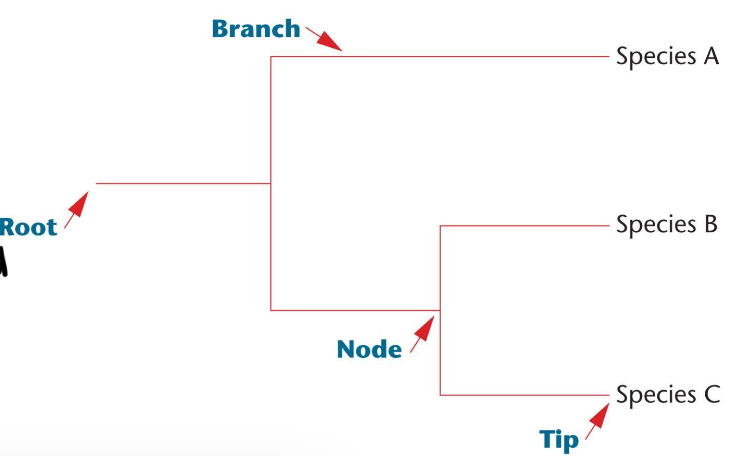
Monophyletic groups
groups consist of an ancestral species an all its descendants
How is genetic variation measured in a laboratory?
genotype individuals’ DNA from a sample population to measure number of individuals with a specific polymorphism
sample population must be random and large enough to accurately represent the entire population
similar techniques to genetics testing and diagnosis, but analyzing many samples at the same time
Examples of techniques
direct DNA sequencing
qPCR
look for DNA marker differences on a gel or blot
qPCR to measure amounts of 2 alleles
amplify locus of interest using primers and primer probes directed against specific polymorphisms (alleles) in your sample population
probes can be differentially labeled with fluorescence
fluorescence from probe would report how much of each allele is present
RFLP analysis
mutation changes sequence so that a restriction site is added or removed from the locus
isolate DNA from sample individuals in the population you are studying
cut with restriction enzyme
analyze fragments on Southern blot
could also PCR amplify the locus, cut the fragment, and analyze the fragments on a gel
Biological fitness
measure if the number of offspring a particular individual/phenotype will produce
Speciation
the process of splitting a genetically homogenous population into 2 or more populations
become reproductively isolated
as mutation occurs, barriers of reproduction will form
Founder mutation
first individual in population to acquire a mutation
Founder effect
one or a few individuals contribute alleles to next generation
decreases variation