09 - Cytoskeleton and Motors: Actin and Myosin
1/83
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
84 Terms
Cytoskeleton
system of protein filaments in the cytosol that give the cell its shape and ability to move in a directed fashion.
Three types of filaments:
• Actin filaments
• Microtubules
• Intermediate filaments
Key characteristics of The Cytoskeleton
• Formed by polymers of proteins (non-covalent bonds)
• Rapid assemble and disassembly required for function
The Cytoskeleton is Involved in:
• Providing structure and support (scaffolding)
• Acting as internal framework to position organelles
• Allowing movement of materials and organelles (intracellular transport)
• Required for cell motility and function
• Machinery for cell division
Actin Filaments (Microfilaments)
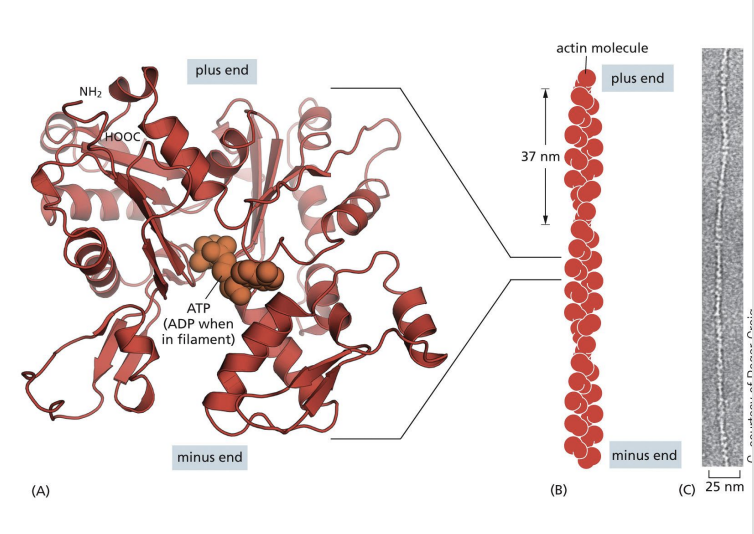
Actin filaments are 8nm diameter filaments formed by monomers, G-actin (globular actin) which polymerizes into F-actin (filamentous actin).
F-actin polymers assemble into double helix (one twist every 37nm) and have polarity
the ends in Actin Filaments (Microfilaments)
(-) end = pointed end
(+) end = barbed end
Actin Filament Nucleation
Nucleation = formation of initial oligomer (acts as a nucleus) which can be used for elongation
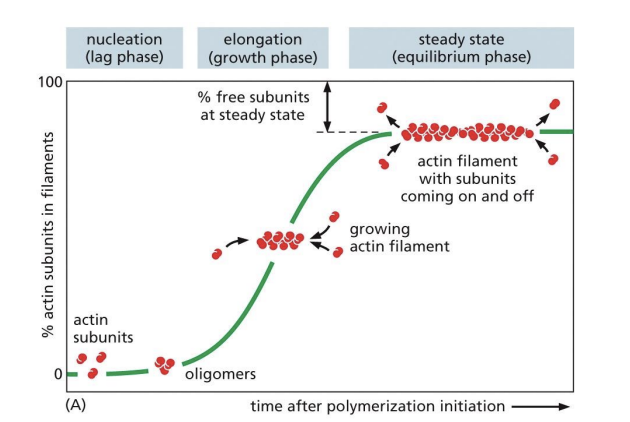
Actin Filament Polymerization
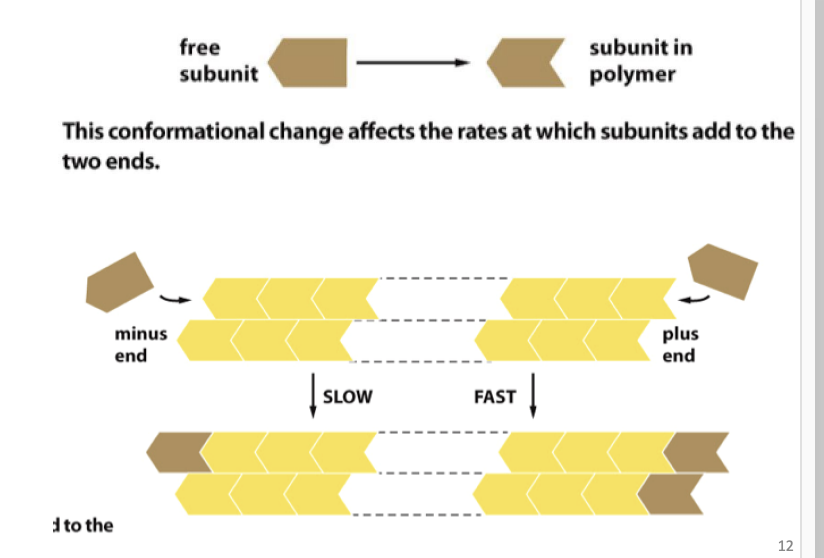
Polymerization can occur at either end (addition of actin monomers to either (-) or (+) ends), if actin monomer concentration is high.
If monomer concentration is low, addition occurs on barbed end (+).
(+) = fast growing
(-) = slow growing
Actin Filament Depolymerization
Depolymerization typically occurs at pointed end (-).
Actin is constantly assembling and disassembling
Actin Polymerization
Polymerization requires energy = ATP
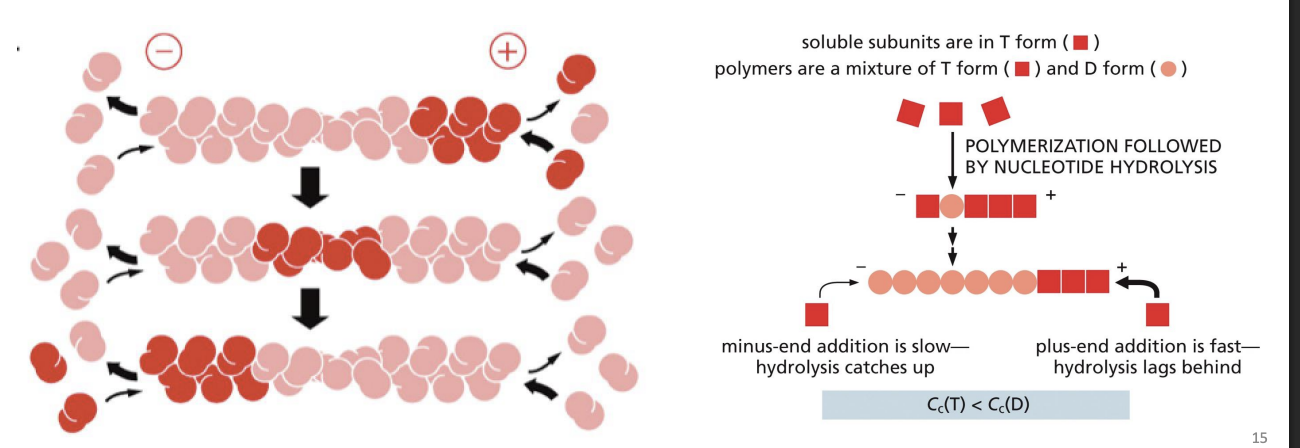
Actin carries tightly bound ATP that is hydrolyzed to ADP soon after assembly into polymer.
Most actin filaments = ADP-Actin subunits
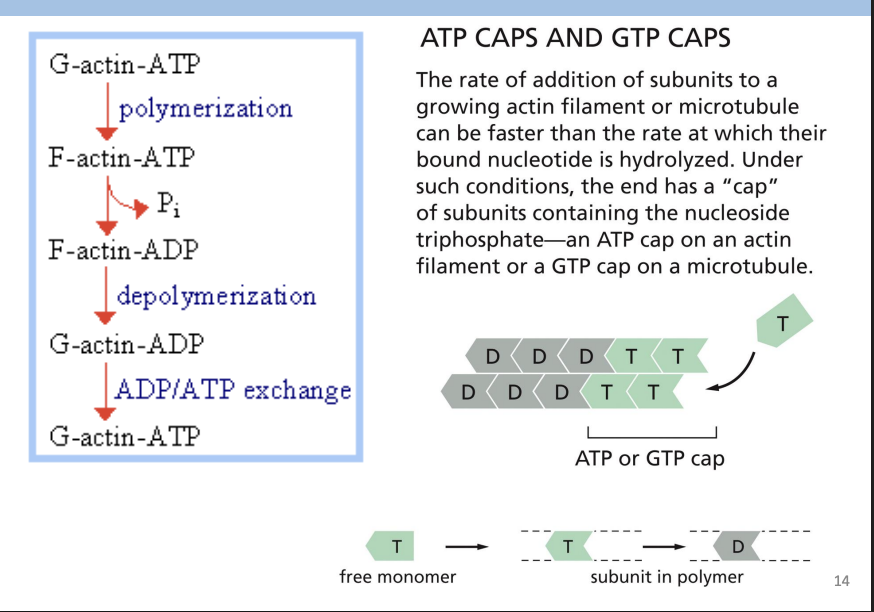
Actin Filament Treadmilling
Monomers being preferentially added to one end (+) and removed from the other (-) = monomers appear to move towards pointed end = “Treadmilling”
types of actin-binding protein
More than 100 actin binding proteins!
some classes are:
Nucleating proteins
Monomer binding proteins
Capping proteins
Nucleating proteins
begin actin polymerization (first step is assembly of 2-3 monomers) e.g. Arp 2/3 complex and formin.
Remains bound to (-) end to allow for rapid elongation on (+) end.
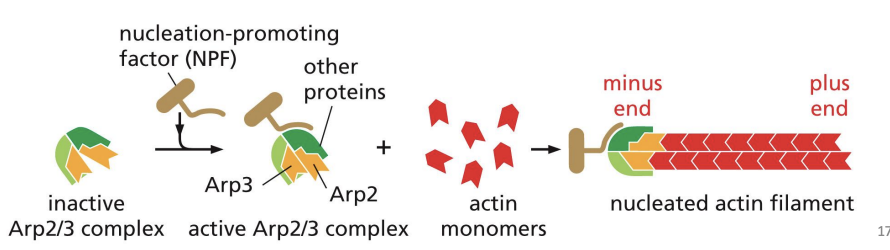
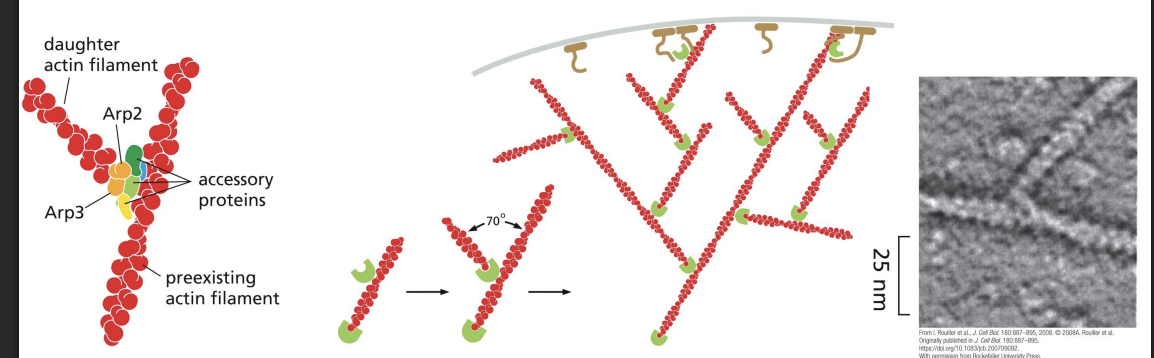
Arp 2/3 complex
nucleates assembly to form a branched network and remains associated with the minus end
Arp2/3 can also bind to pre-existing actin filaments, and is most efficient at nucleating these sites, creating branches at a 70o angle.
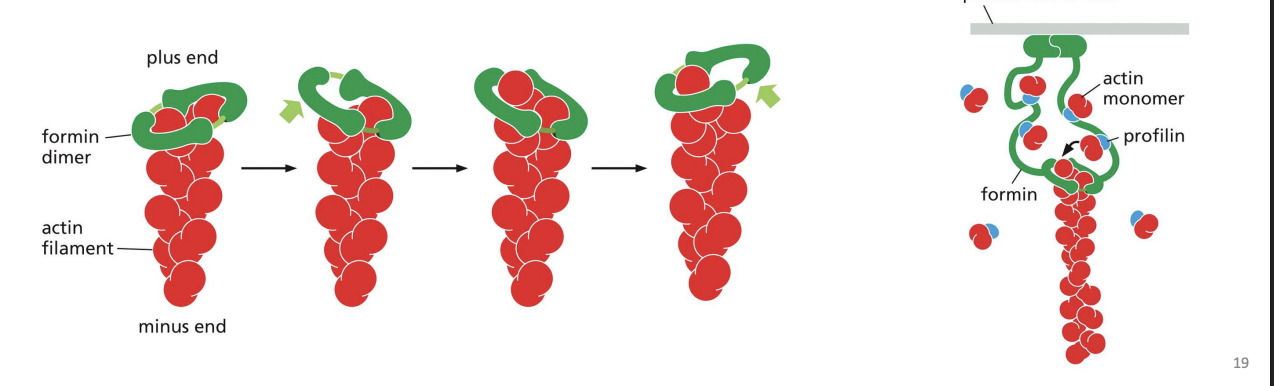
Formins
dimeric proteins that can nucleate growth of unbranched filaments to cross-link and form parallel bundles. Formins do not block the end that they are associated with (+ end), preventing loss of subunits from that end.
Monomer binding proteins
bind Gactin (ATP-Actin) to either prevent polymerization (e.g. Thymosin) or promote it (e.g. Profilin).
Profilin
protein that binds to actin monomers opposite the ATP binding cleft, blocking the side that interacts with the (-) end of a filament (leaves only (+) end binding site free). Profilin falls off when the actin subunit binds to an actin filament.
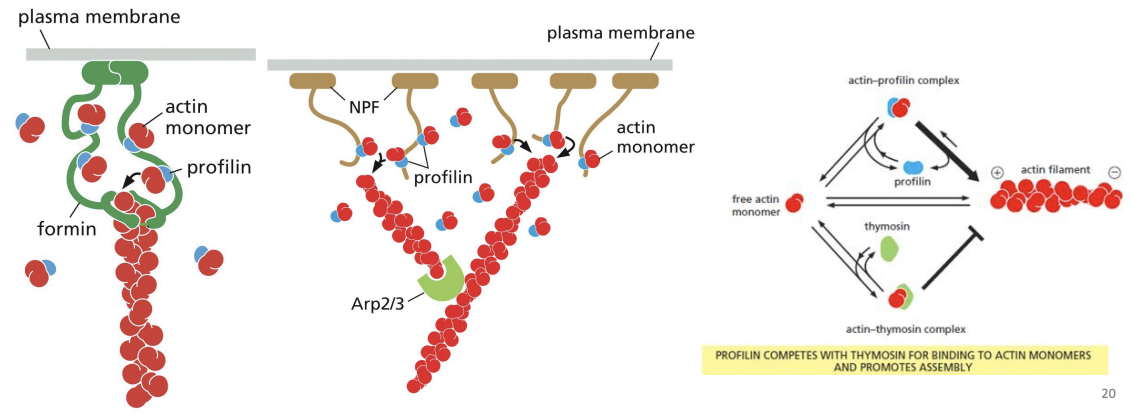
Thymosin
binds subunit prevents asesembly
Capping proteins
bind + of F-actin to prevent growth or shrinkage (regulates length of filaments)
Uncapped filaments would depolymerize rapidly.
prevents assembly and disassembly at plus end
Critical concentration
rate of subunit growth is equal to rate of subunit loss.
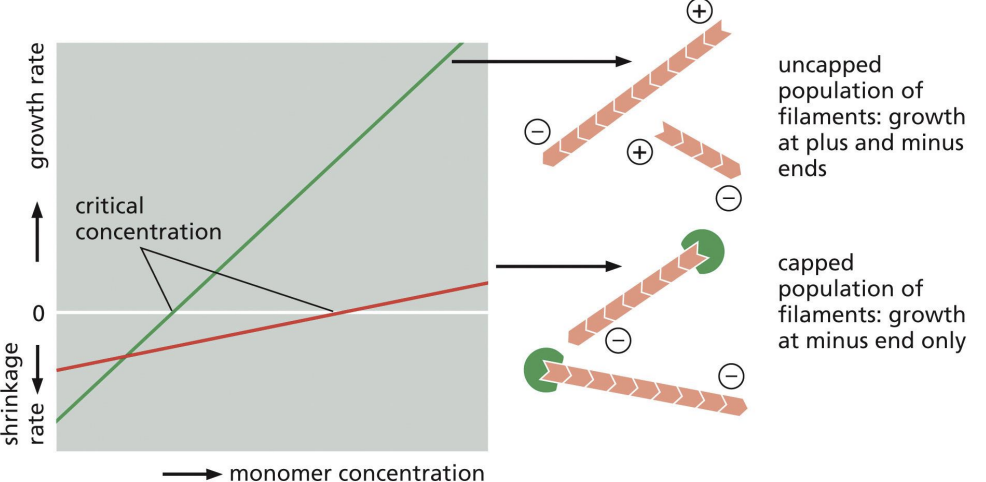
Tropomodulin
bind – end of F-actin to prevent growth or shrinkage (regulates length of filaments)
prevents assembly and disassembly at minus end
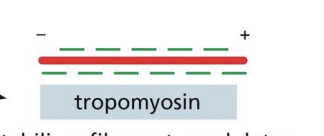
Side-binding proteins
bind multiple actin subunits to stabilize and stiffen the filament and prevent other interactions (e.g. Tropomyosin)
Tropomyosin
stabilizes filament, modulates binding of other accessory proteins
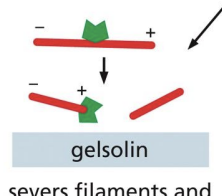
Severing/De-stabilizing proteins
= proteins that break actin filaments into smaller filaments (e.g. Gelsolin which caps end after severing and Cofilin which forces twisting of filament to sever it (less table)
Gelsolin
servers filament and binds to plus end
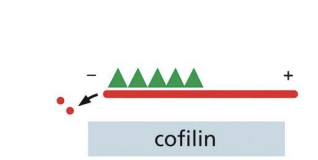
Cofilin
protein that twists actin filament tighter to create mechanical stress that weakens the contacts between subunits, making it less stable and easily severed by thermal motions.
Tends to bind ActinADP subunits (D state).
Cross-linking proteins
Direct organization of microfilaments into stable network (bundling proteins) adding rigidity/support in cell e.g. Fimbrin (tight packing), alpha-actinin (loose packing) and Filimin (3D web)
Plasma membrane binding proteins
Bind actin filaments and connect to PM, required for cell motility e.g. Spectrin family, Dystrophin, ERM
why do most accessory proteins bind to filaments
most accessory proteins bind to filaments to determine spatial distribution and dynamic behaviour
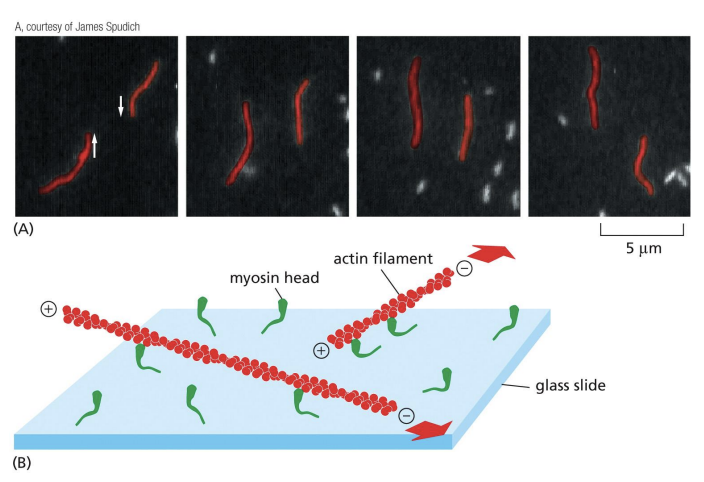
Motor proteins
proteins that bind to a cytoskeletal filament and use energy derived from repeated cycles of ATP hydrolysis to move along it.
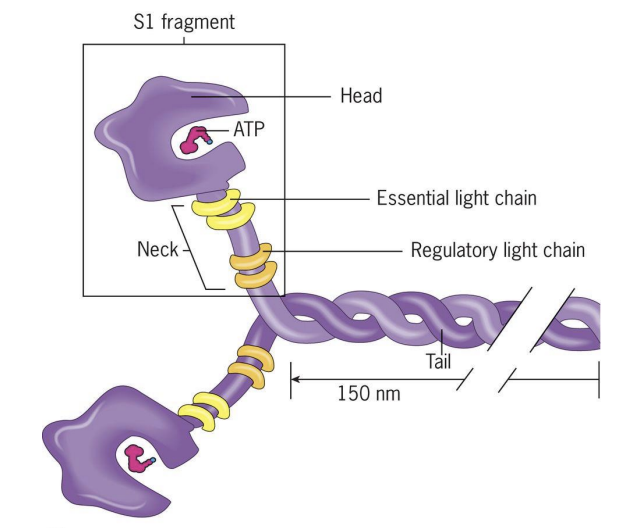
Myosins
Motors that move along F-actin towards the barbed (+) end and use ATP to drive movement.
Composed of 1 pair of heavy chains and 2 pairs of light chains.
Heavy chain has “head domain” = ATPhydrolysis domain (power generation) and actin binding region.
2 classes of myosin
2 classes =
1) Conventional myosin (E.g. Myosin II) and
2) Unconventional Myosins (at least 17 classes e.g. Myosin V)
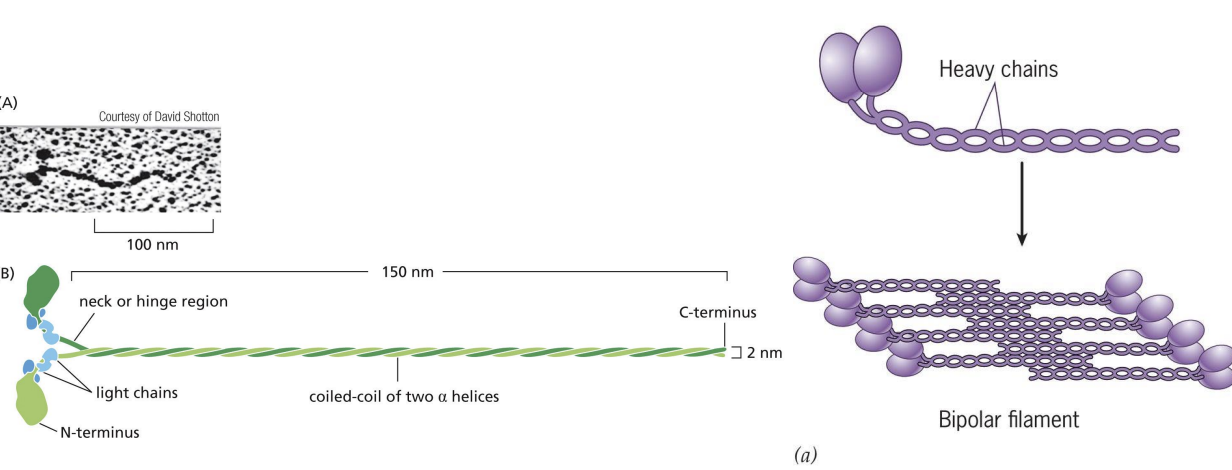
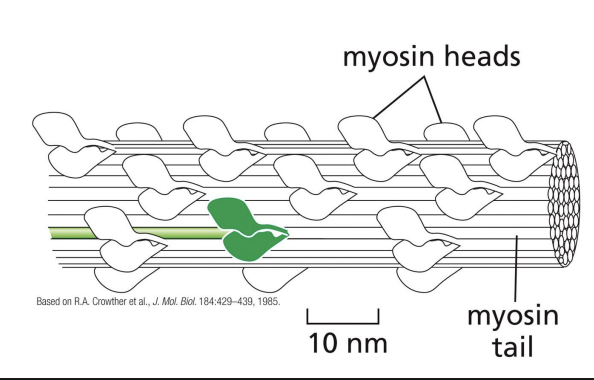
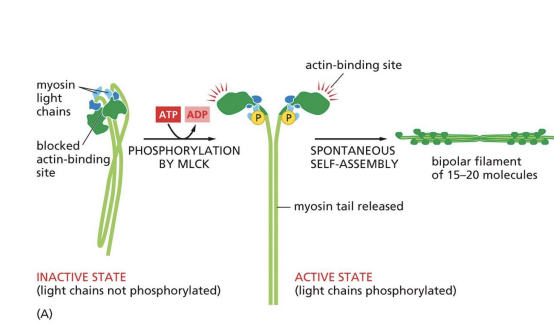
bipolar filament
Heavy chains coil together to form “tail” = protein forms filaments (bipolar filaments) which staggers arrangement of myosin molecules, so tails point inwards and heads point outwards
Bipolar filament structure
Bipolar filament has a central bare zone = free of myosin head domains.
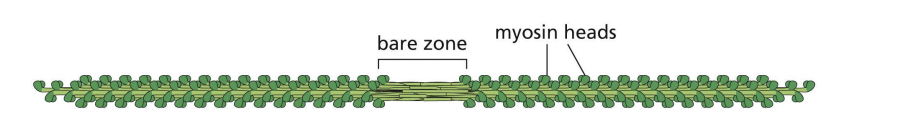
resting state of myosin
In resting state (non-contracting), two myosin heads are bent backwards and sterically interfere with each other (activity = off)
Myosin Motor Protein Activity
Each myosin head binds and hydrolyzes ATP, using energy to walk towards (+) end of actin filament.
Opposing orientation of myosin heads in bipolar filament = efficient at sliding pairs of opposite orientation actin filaments towards each other
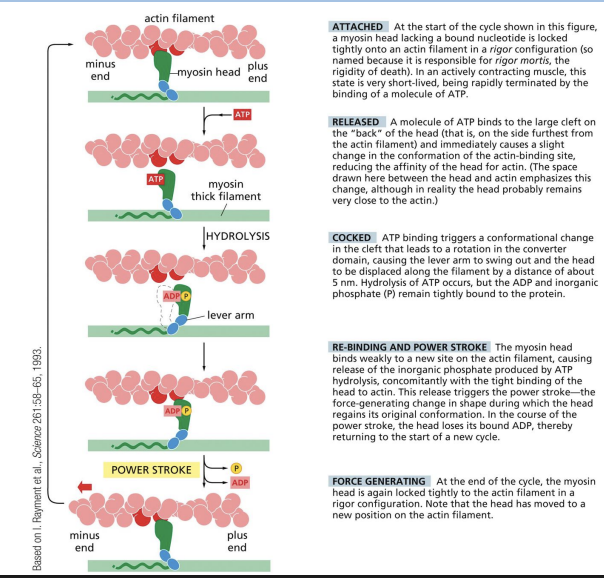
ATP-driven actin–myosin motor cycle
Each cycle of ATP binding, hydrolysis and release = propelling of motor protein forward to new binding site.
1) ATP binding to myosin head releases the myosin head from the actin filament.
2) ATP hydrolysis and rotation of myosin head (swinging of 8.5nm lever arm).
3) Binding of myosin head to actin filament.
4) Release of phosphate group and “power stroke” = force generating change restoring myosin head to original conformation = single step of movement.
5) Release of ADP from myosin head.
Role of Myosin in Muscle Contraction
Myosin II required in most cells for cell motility and cytokinesis during cell division, muscle cells require it for muscle contraction
Muscle cells =
very long, cylindrical cells with hundreds of nuclei = muscle fibers (formed by fusion of several smaller cells)
Muscle fibers =
collection of myofibrils.
Myofibrils =
organized repeating structures (sarcomeres).
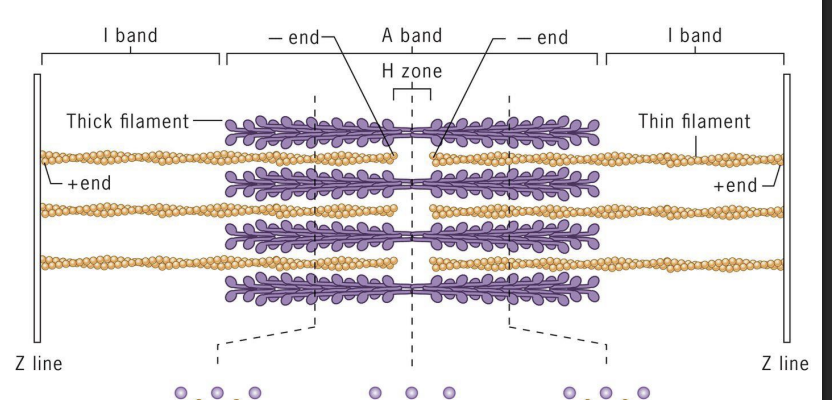
Sarcomeres
array of precisely ordered parallel thick and thin filaments with overlap between them (contractile units of muscle).
Thin = actin filaments (microfilaments)
Thick = myosin II bipolar filaments
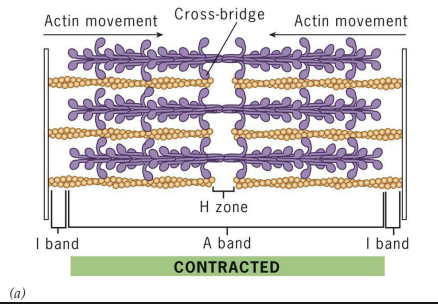
Actin and Myosin in Muscle Contraction
Actin (+) ends attached to Z discs at end of each sarcomere.
M line (midline) = location of proteins linking adjacent Myosin II proteins.
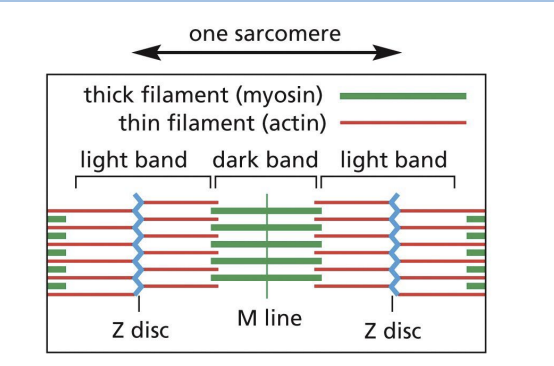
Sarcomere contraction
actin and myosin slide past each other without shortening.
I-band =
thin (actin) filaments only.
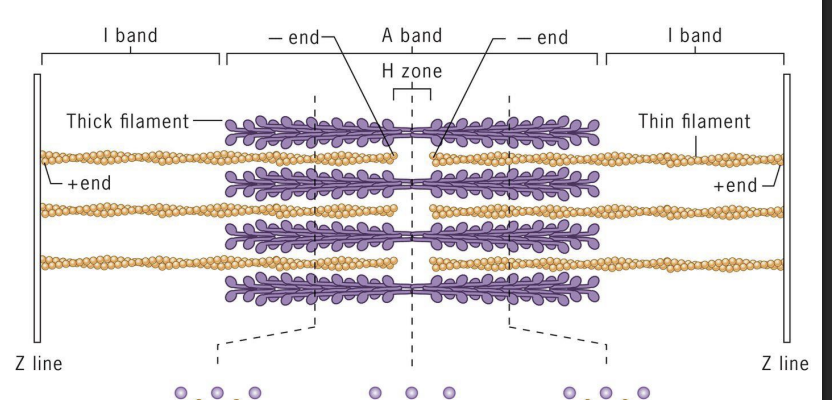
H-zone =
central region of A-band with only thick (myosin) filaments.
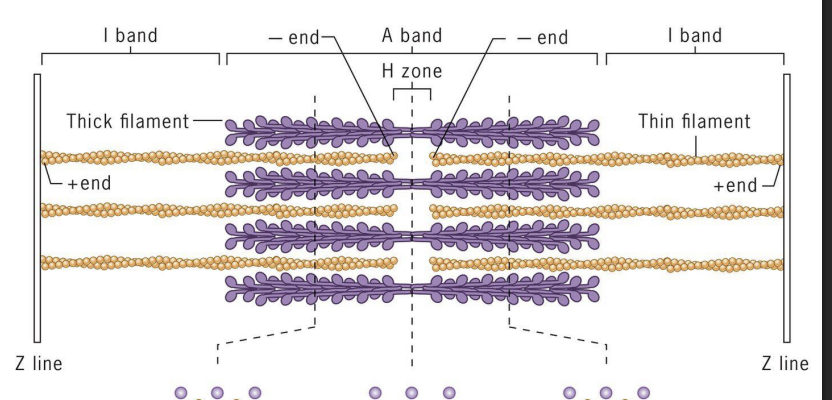
A band =
region of thick filaments and overlapping thin filaments
Relaxed state in Muscle Contraction
sarcomere at maximum length, has wider I-bands and H-zone (no cross-bridges).
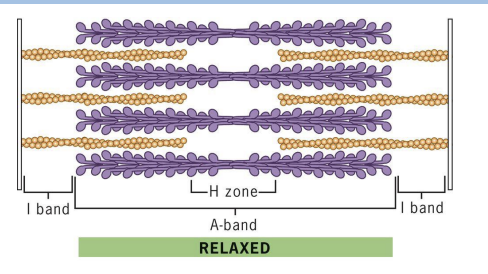
Contracted state in Muscle Contraction
Sarcomere with reduced length (filament lengths don’t change). I-bands and H-zone greatly reduced. Actin filaments are moved by myosin crossbridges
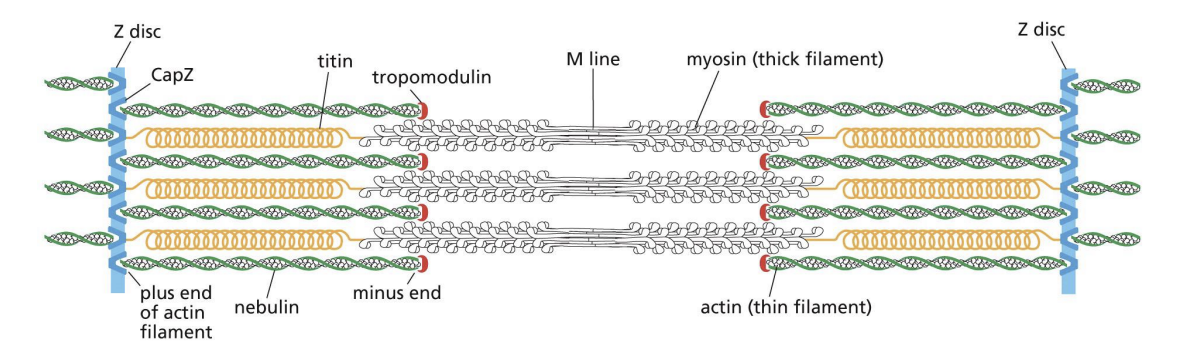
Accessory Proteins in Sarcomere
Z disc is built with capping protein (CapZ) and bundling protein alpha-actinin. Tropomyosin and Nebulin stabilize filaments through binding them on the side. Titin = positions thick filaments midway between Z-discs (acts like a spring and changes length as sarcomere contracts and relaxes).
Myosin II bipolar thick filaments slide actin filaments toward one another to bring
about muscle contraction. What arrangement of actin filaments is necessary for
myosin to bring about contraction?
A) An actin filament minus end must face an actin plus end.
B) The actin filament minus ends must face one another.
C) The actin filament plus ends must face one another.
D) The arrangement of plus and minus ends is irrelevant.
c
T-tubules
membranous folds that extend inward from PM around each myofibril.
Action potential triggers release of calcium (Ca2+), which triggers muscle contraction
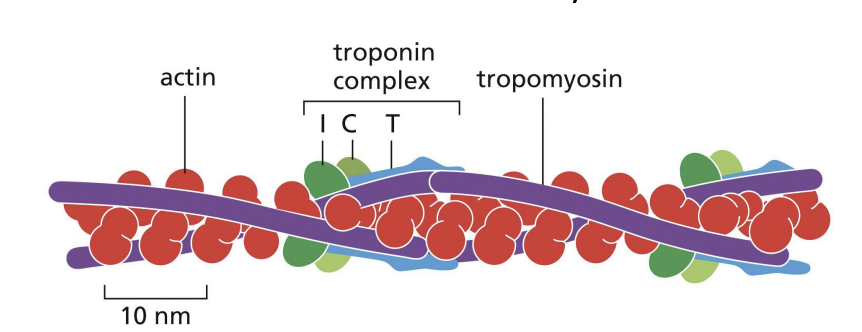
Troponin (Ca2+)
binding protein is attached to tropomyosin.
Ca2+ binding to troponin changes tropomyosin conformation = moves away from actin (myosin heads can make contact with actin)
Increases in Cytosolic Calcium Concentrations Cause
Muscle Contraction
Which of the following would increase the level of muscle contraction? Select all
that apply.
A) Mutation in Troponin such that it no longer binds Tropomyosin.
B) Addition of a molecule to bind free Ca2+
C) Addition of a leaky Ca2+ channel to the sarcoplasmic reticulum.
D) Blockage of the Ca2+ pump.
c and d
Myosin II in Non-Muscle Cells
Myosin II required in most cells for cell motility and cytokinesis during cell division, also involved in stress fiber formation = connections to extracellular matrix
Unconventional Myosin proteins
family of myosin proteins that have either one head or two and are involved in intracellular transport (can bind to other subunits and interact with cargo).
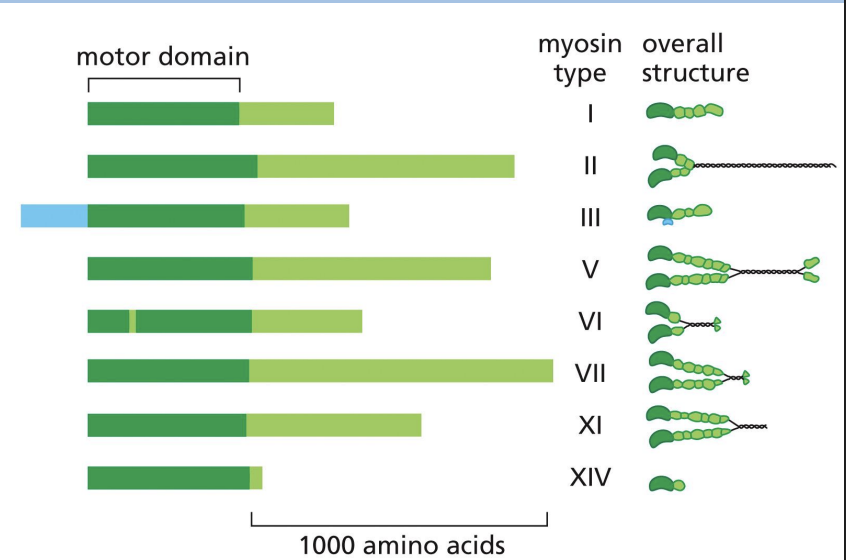
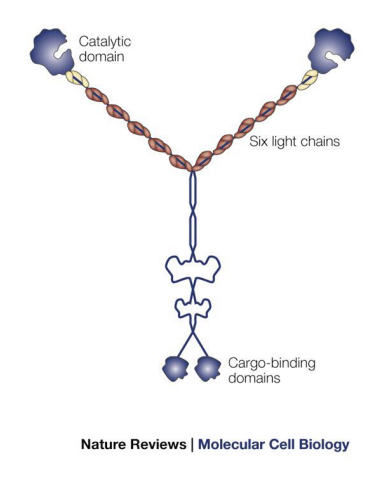
Unconventional Myosin V
2 heads like Myosin II and has very long “neck” region (can make long strides on actin without letting go).
Functions in intracellular transport = head binds actin, tail binds vesicles/cargo, uses ATP to move cargo to (+) end.
“Hand over hand” model: Myosin V
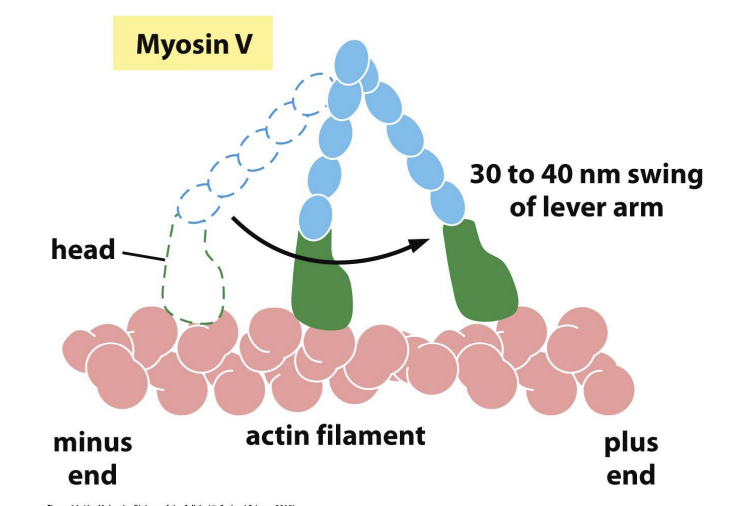
function of Myosin V
carries many kinds of vesicular cargo
Cargo traveling to synapse regions (vesicles) are transferred from microtubule motors to Myosin V
Controls portioning of organelles to daughter cells (mitochondria, peroxisomes).
Microfilaments in Cell Motility
Non-Muscle motility is essential for: development of an embryo, formation of blood vessels, wound healing, movement of immune cells
Spread of cancerous tumour cells
Cell cortex =
specialized, thin layer for cytoplasm just under the PM.
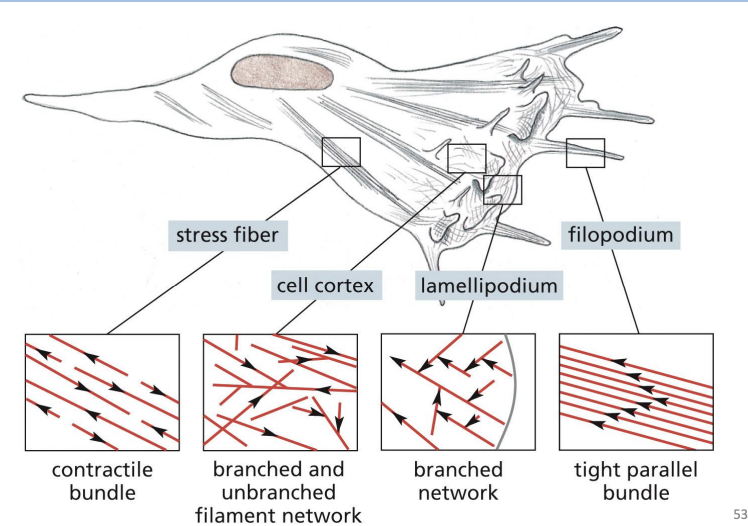
Filopodia =
Thin, spike-like protrusion with actin filament core.
are one-dimensional and help sense environmental cues and aid in cell migration.
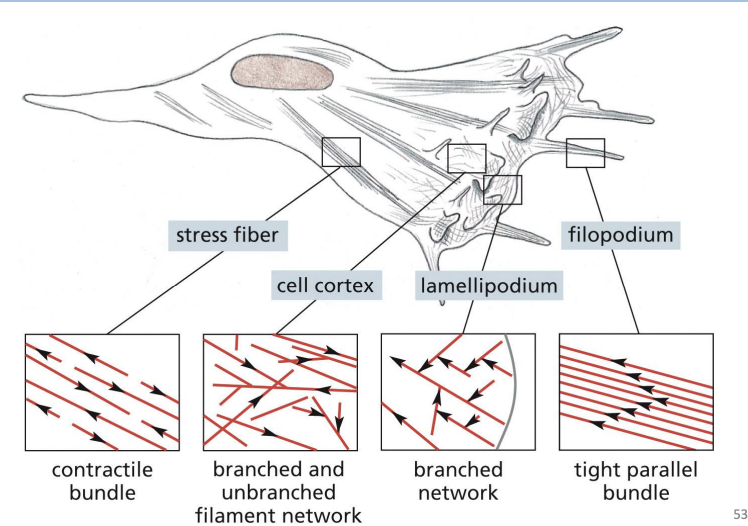
Lamellipodia =
Flattened, sheet-like protrusion supported by meshwork of actin filaments
drive forward movement of migrating cells
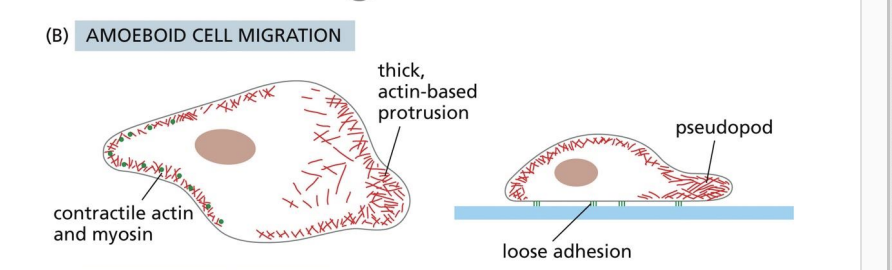
Pseudopodia
3D protrusions that are thicker than lamellipodia.
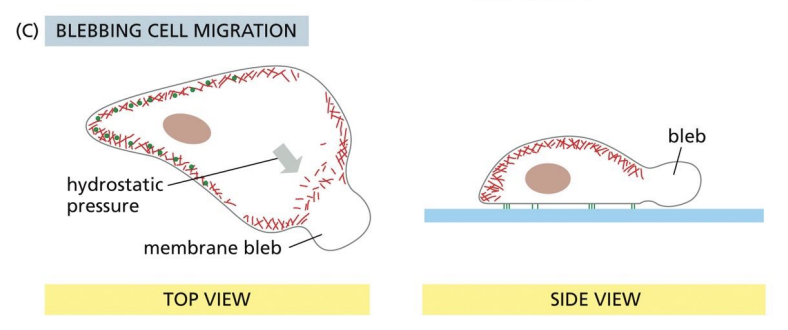
Blebbing
3D protrusion where PM detaches from actin, cytoplasm pushes membrane forward.
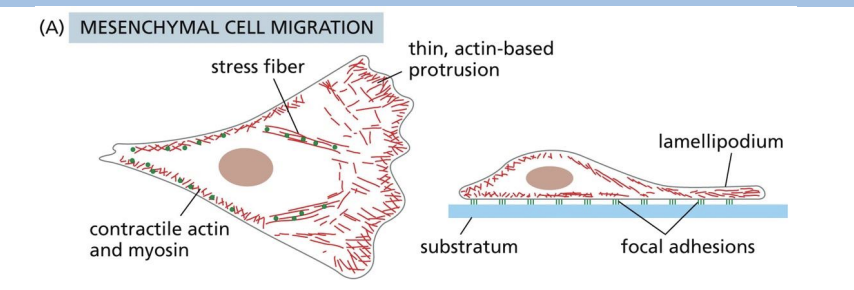
mesenchymal crawling
Filopodia + Lamellipodia
amoeboid crawling
Pseudopodia
blebbing migration
Blebs
Which one of the following arrays of actin filaments is associated with filopodia?
A) Branched and unbranched filament network
B) Tight parallel bundle
C) Contractile bundle
D) Branched network
b
Microfilaments in Cell Migration
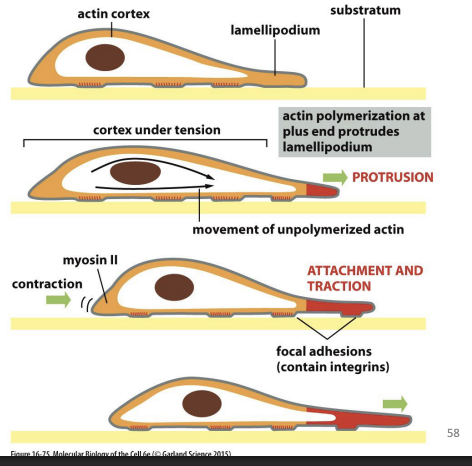
Series of repetitive actions:
1) Cell protrudes in direction it wants to move (lamellipodium)
2) Cell protrusions attach to surface beneath
3) Bulk of cell is pulled forward towards surface contacts
Cell protrusion/forward pulling forces generated by actin polymerization.
Retraction forces in rear of cell is generated by Myosin II.
Microfilaments Direct Cell Motility
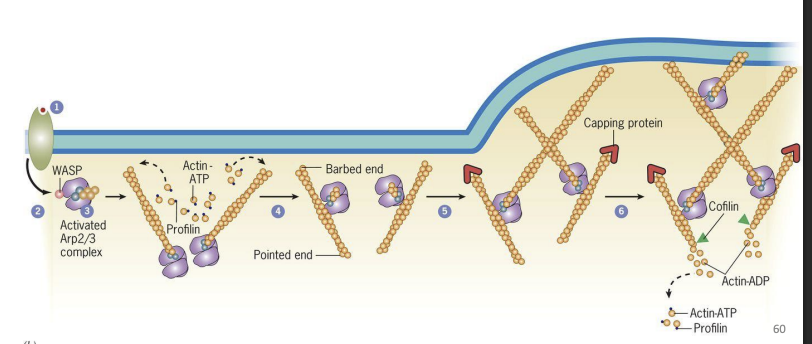
1) Stimulus received on one end of cell (PM receptors bind to proteins)
2) Family of proteins in cytosol get activated = WASP (Wiskott-Aldrich Syndrome) proteins
3) Activated WASP proteins activate Arp2/3 complex proteins
4) Activated Arp 2/3 proteins serve as nucleating site for new actin filaments (new actin filaments form)
5) Activated Arp2/3 proteins bind to new actin filaments and nucleate more
filaments (branches)
6) Growing barbed (+) end push PM outward in direction of stimulus
As newer actin filaments form, and older filaments are disassembled by proteins like Cofilin
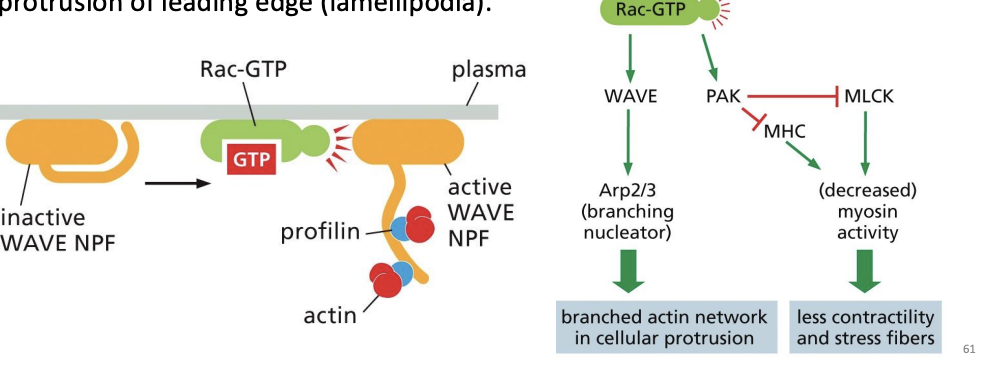
What role do Rac-GTP and WAVE proteins play in microfilament organization during cell migration?
Once polarity is established by cdc42 proteins, Rac-GTP and WAVE proteins stimulate Arc2/3 proteins to nucleate branched actin filaments and drive protrusion of leading edge (lamellipodia).
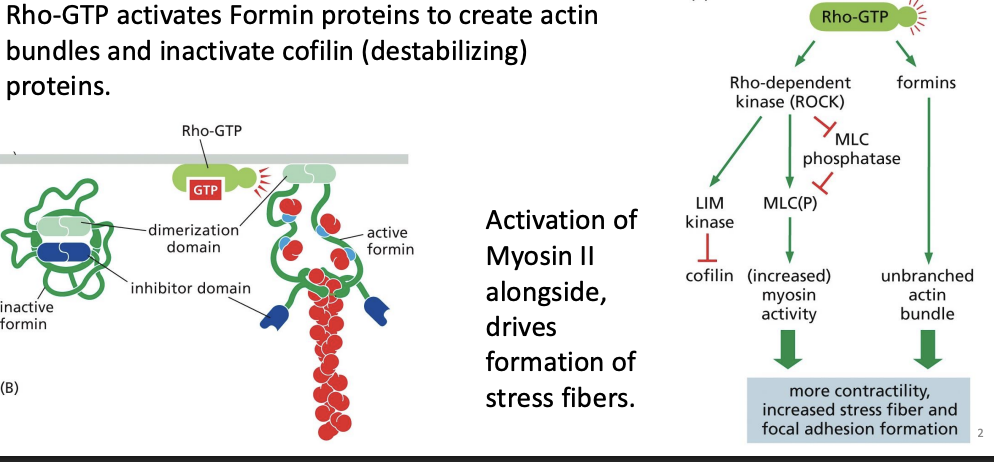
What role does Rho-GTP play in microfilament organization during cell migration?
Rho-GTP activates Formin proteins to create actin bundles and inactivate cofilin (destabilizing) proteins.
Activation of Myosin II alongside, drives formation of stress fibers
Activation of the monomeric GTPase Rac leads to alterations in actin accessory
proteins that promote the formation of protrusive actin networks in lamellipodia
and pseudopodia. Which one of the following statements correctly describes a
molecular consequence of the activation of WAVE and PAK by Rac-GTP?
A) Rac-GTP activates myosin and increases membrane contractility.
B) Rac-GTP stimulates the formation of unbranched actin filaments.
C) Rac-GTP increases branched actin networks in cellular protrusions.
D) Rac-GTP increases the number of stress fibers throughout the cell
c
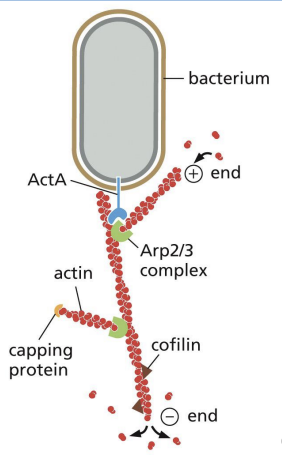
Actin-Based Motility of Pathogens (e.g., Listeria)
Bacteria can hijack host cytoskeleton (e.g. Listeria monocytogenes)!
ActA on bacteria surface activates Arp2/3 and allows for polymerization of actin tails at rear surface of bacteria, propelling it forward = actin “comet
How does Salmonella invade host cells
Salmonella = attaches to epithelial cells, injects proteins that activate WASP and/or Arp2/3 proteins.
Induce lamellipodia formation and membrane ruffles, which causes cell to engulf Salmonella.
How does Enteropathogenic E. coli (EPEC) manipulate host actin?
Enteropathogenic E.Coli = binds epithelial cells of intestine.
Injects proteins that activate WASP and/or Arp2/3 proteins and causes polymerization of actin (formation of actin pedestals).
Normal microvilli disappear = poor absorption of nutrients.
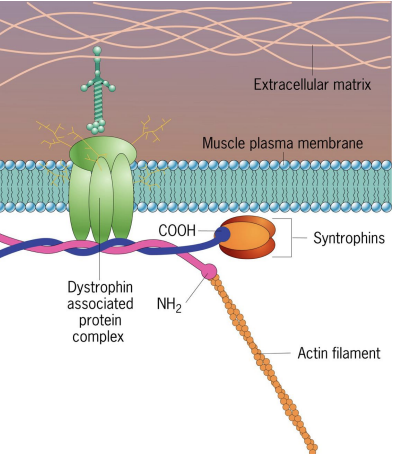
Duchenne-type Muscular Dystrophy (DMD)
DMD = Lack of protein, Dystrophin (anchor between actin filaments and PM in muscle cells), causes lack of structural support.
Affects both cardiac and skeletal muscle leading to deterioration
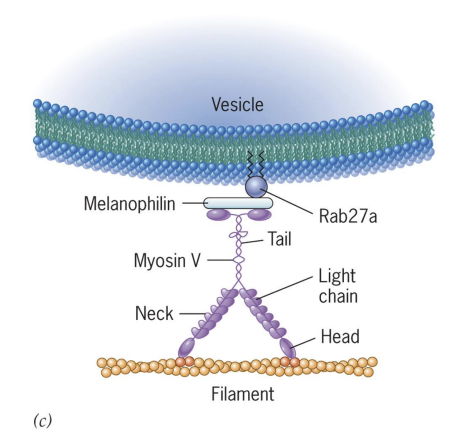
Griscelli Syndrome
Griscelli syndrome = Humans lack unconventional Myosin Va
Myosin Va required to transport pigment granules (melanosomes) in pigment cells (melanocytes).
Melanosomes are moved to cell periphery to be transferred to and incorporated into developing hair = albinism due to lack of transport.
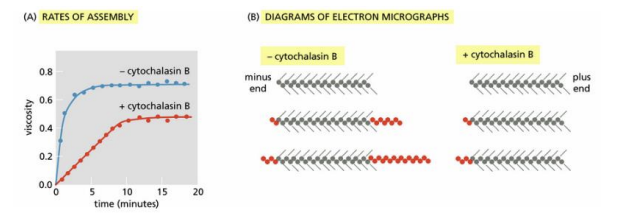
Cytochalasin B interferes with the assembly of actin filaments. In the classic experiment that defined its mechanism, short lengths of actin filaments were decorated with myosin heads and then mixed with actin subunits in the presence or absence of cytochalasin B. Assembly of actin filaments was measured by assaying the viscosity of the solution (see the figure Part A) and by examining samples by electron microscopy (Part B). The decorated actin filaments present before the addition of actin monomers are shown at the top of each set of three. Filaments present after increasing times of incubation with actin monomers (red circles) are shown below
Which of the following statements describes a plausible mechanism of cytochalasin B action that accounts for the slower rate of assembly and the appearance of the filaments in the electron micrographs?
A) Cytochalasin B binds to the plus end of the filament to block monomer addition.
B) Cytochalasin B binds to the actin monomers to promote addition to the plus end.
C) Cytochalasin B binds to the actin monomers to promote addition to the minus end.
D) Cytochalasin B binds to the minus end of the filament to block monomer addition
a
Most eukaryotic cells maintain the concentration of actin monomers well above
the critical concentration needed for actin polymerization in the test tube. In most
cells, the excess actin monomers are bound by the protein profilin, which
regulates their addition to actin filaments. Why is this strategy advantageous for
the cell?
A) Profilin lowers the critical concentration of actin so filaments can grow.
B) Profilin binding ensures that the cell will not run out of actin monomers.
C) Profilin prevents cells from making excessive numbers of actin filaments.
D) Profilin allows actin monomers to be recruited to sites of filament growth
d
Some actin-binding accessory proteins significantly increase the rate at which the
formation of actin filaments is initiated in the cytosol. How might such proteins do
this? What must they not do when binding the actin monomers?
A) They promote polymerization by destabilizing complexes of monomers; they
must not block the ends required for polymerization.
B) They promote polymerization by stabilizing complexes of monomers; they
must not block the ends required for polymerization.
C) They promote polymerization by stabilizing complexes of monomers; they
must not bind to the sides of the actin monomers.
D) They promote polymerization by destabilizing complexes of monomers; they
must not bind to the sides of the actin monomers.
b
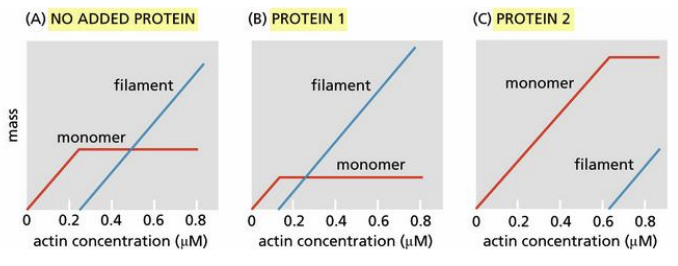
You have two proteins that you suspect cap the ends of actin filaments. To determine whether they do and, if so, which protein caps which end, you measure filament formation as a function of actin concentration in the absence of either protein, in the presence of protein 1, and in the presence of protein 2 (see the figure). The mass of actin, as monomers or filaments, was determined at equilibrium
Which end of the actin filament is capped by protein 1? Which by protein 2?
A) Protein 1 caps the plus end; protein 2 caps the minus end.
B) Protein 1 caps the minus end; protein 2 caps the plus end.
C) Protein 1 caps the plus end; protein 2 caps the plus end.
D) Protein 1 caps the minus end; protein 2 caps the minus end.
b