BIOL 325 DNA POLYMORPHISMS STUDY GUIDE
1/94
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
95 Terms
what is a gene polymorphism?
a sequence difference that is present in at least 1-2% of the population
can be single bases (SNP) or thousands of bases
may or may not have phenotypic effects
are the reason for alleles - gene variants
found throughout the genome but is more common in some areas than others
what is a single nucleotide polymorphism (SNP)?
genome sequences that differ by at least one NT every 1,000-2,000 bases
if the location of a polymorphic sequence what can it be served as?
it can serve as a landmark or marker for locating other genes or genetics regions
what are each polymorphic markers that have different versions called? what is their alternative name and why?
they are called alleles
alternative name = morphs because alleles are usually used for expression terms
what is the HLA locus?
REALLY polymorphic and creates the WBC
what percentage are unique DNA sequences
about 1% of our genes
what are repeated DNA sequences broken up into? what do each category contain?
tandem repeats (side by side repeats) and dispersed repeats (repeats in different spots)
tandem repeats contain tandem paralogues, satellite DNA (satellites, mini, and micro), and rDNA
dispersed repeats contain transposons which are jumping genes
what is the 1000 genomes project? what was the conclusion? what is ok to have?
sequenced 1000 genomes of different people so it is diverse and looked at how many SNPs there are
concluded that SNPs are common and are mostly located at the ends of the genome
it is ok to have SNPs at the ends because the ends are mostly lost in replication and the most important egens are not at the ends
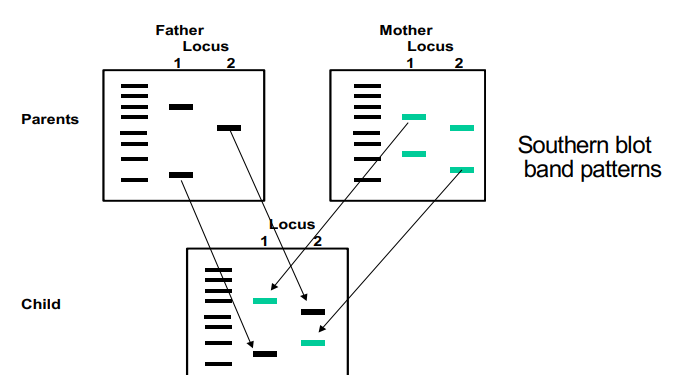
define RFLP, the technique to distinguish RFLP, and the situations where you might want to detect RFLPs (written response question)
RFLP’s are restriction fragment length polymorphisms and are distinguished by southern blotting through the use of restriction enzymes. The restriction fragment sizes are altered by changes in or between enzyme recognition sites and single nucleotide polymorphisms change the restriction enzyme sites. The situations where RFLPs may be detected is to identify a person, identify the father of a child by comparing the father’s loci to the child’s loci, and to exclude or establish a suspect is tied to a crime scene.
what is the process for detecting RFLPs? what will a mutant, heterozygous, and wild type look like on a gel?
a long sequence that is PCR’d should have two restriction enzyme sites for whatever the enzyme is
one will cut both wild type and mutation and two is at the site of the mutation
two will cut in some situations but not in others
both alleles are wild type (+/+) = will cut twice and get three bands
both the alleles are mutants (-/-) = neither of the cut sites will work and will be one band on the gel
alleles are homozygous (+/- or -/+) = one fragment doesn’t cut in one location and the other fragment cuts in another location (either at the mutant or at the first location) and will have two bands on the gel
how are RFLPs used to determine who the father is for a child?
RFLPs are inherited so for each locus, one allele is inherited from each parent
on a southern blot or simply on a gel, one locus will match the father’s RFLP and the mother’s RFLP on the same locus
one difference in just ONE allele excludes paternity in that father
what is the standard reference material (SRM) DNA profiling list for RFLP analysis good for?
good for identification like determine who the father/parent is for a child
how can RFLPs be used to test evidence?
a suspects RFLPs will correspond to the evidence bands which can exclude or include if that suspect was at the crime scene
to see who the suspect is, substract out the victims bands that correspond to the evidence from the crime scene then compare the two suspects bands to the evidence from the crime scene
what is exclusion easier? why is inclusion harder?
for exclusion = based on the data, you can just exclude a suspect because they do not match
for inclusion = nothing in science is 100% which can only determine probability
define STR, the technique to distinguish STR, and the situations where you might want to detect STRs (written response question)
STR’s are short tandem repeat polymorphisms that repeat nucleotide sequences less than 8 times. What is polymorphic in STR’s is how many times the nucleotides/nucleotide sequences are repeated and the number of repeats is specific to an individual. The technique that is commonly done to distinguish STRs is through PCR and analyzed through electrophoresis like capillary gel electrophoresis with the chromatograph. STR alleles can be analyzed by their amplicon size through PCR. The situations where STRs may be used are to identify human remains, to establish or exclude paternity, or to match a suspect to a crime scene sample.
how is multiplex PCR used for STR? give an example
multiplex PCR is used to genotype multiple loci in the same reaction
ex.
one allele has the repeat and PCR is used to figure out how many times that repeat is repeated
another allele might have a different sample of repeats but has the same repeated sequence as the first allele
what are the disadvantages of using southern blotting for STRs? what are the advantages of using PCR for STRs?
disadvantages
requires 0.1 to 1 microgram of DNA which is hard to get that much from a crime scene
large fragile gels are used which are time consuming and can break
radiation (32P) is used
advantages
less sample required
faster
cheaper
more sensitive once optimized

what was the example in class for STRs?
allele 1 has 7 repeats making PCR amplicon that has 187 base pairs
allele 2 has 8 repeats making PCR amplicon that has 191 base pairs
they have a 4 base pair difference between the alleles making a four nucleotide repeat for STR individual units
where should primers be for STRs? where should primers NOT be at?
primers must FLANK outside the STR element because they can pick up sequences that aren’t STR which makes the amplicon bigger
primers should NOT be at the middle because they can prime other locations
what are allelic ladders used for in STR’s? what do ladders indicate?
standards representing all alleles in a population
can be purchased for different STRs that are constantly sequenced
are always on both sides of a gel
ladders indicate the number of repeats in the individuals two alleles
what direction are STRs read on a gel? what about through capillary electrophoresis? what do the peaks do in capillary electrophoresis?
STRs are read from bottom to top when using gel electrophoresis because smaller fragments will run through the gel faster, while bigger fragments will stay near the well
STRs are read from left to right on the x axis when using capillary electrophoresis because smaller STR will show up first/closer to the detector
the peaks in capillary electrophoresis will be well optimized to clearly see the number of repeats and will show the amount/signal amplitude of the repats
how are STR genotypes read in a chart?
since STR genotypes are inherited and can be used to see who the father is for a child,
the peaks in the child will match to the peaks from the father and mother at the same location for each peak
if a peak or peaks aren’t clear because sometimes they aren’t working well = not detectable/analyzable
a good sequence would get all 13 loci
define VNTR, the technique to distinguish VNTR, and the situations where you might want to detect VNTRs (written response question)
VNTRs are variable number of tandem repeats that are 8 to >50 base pair minisatellites. The technique that can be used to distinguish VNTR is through microassays like southern blotting and if the repeats are in the 100s or are smaller, PCR can be used. The situations where a researcher might want to detect VNTR is to identify the severity of a disease that is correlated with variation of STR number like Huntington disease. A parent with Huntingtons can test their child relating to their VNTRs to get an idea of what age they will start presenting symptoms. More repeats means earlier symptoms and the worse the symptoms will be.
what are the other diseases that can be seen with VNTR?
fragile X
myotonic dystrophy
friedreich ataxia
kennedy disease
haw river syndrome
spinocerebellar ataxia
what are microvariants?
STRs that occasionally contain repeat units with altered sequences
what is the STR nomenclature for genes? give an example
first two letters means the gene short handed
the end is the number for the intron
ex. TH01 = intron 1 of tyrosine hydroxylase gene chromosome 11
what is the STR nomenclature for non-genes? give an example
uses the D#S# system which indicates chromosome number and segment number
ex. D21S11 is chromosome 21, segment 11
what are the desirable features for STRs?
high heterozygosity = they are unique within the population because most of our DNA is not that different from each other
regular repeat unit = it always has the same repeat like the same trinucleotide repeat
distinguishable alleles = a great primer and probe system can distinguish alleles from each other
robust amplification = do not have a lot of DNA → PCR will amplify the sample
what is the STR nomenclature for microvariant alleles with partial repeats? give an example
number of complete repeats followed by a decimal point before partial repeats
ex. 9.4
what did the FBI adopt in 1997? what does it have? what is D3S1358?
adopted the 13 “core” loci as the Combined DNA Indexing system, CODIS which is still used today
has genes and non genes
DS3158 = a particular segment in heterochromatic DNA that is highly variable
what was the article, Forensic Autosomal STR and Their Potential Association With Phenotype?
was more of a recent use of STR (2022)
trying to use STR to try to associate those genotypes with phenotypes like schizophrenia
more repeats = worse schizophrenia
also looked for biomarkers and their repeats for gastric cancer patients and if parents has had gastric cancer then would test for repeats in children
what is HUMAMEL?
it is human amel and codes for amelogenin-like protein (makes enamel) and is not an STR
the gene is located at Xp22.1-22.3 and Y
not used to see how different we are from a woman or a man, used to see if we are a woman or a man
what tests are most commonly used for sex determination? (written response question)
One test that can be done for sex determination is the amelogenin sex test. In this test, the ameologenin gene on both the X and Y chromosomes are PCR amplified and resolved on a gel. The female would have one band since only AMLEX is present and a male would have two bands because AMLEX and AMELY are present.
how many base pairs in the X allele? what about the Y allele? what are females and males?
X allele = 212 base pairs
Y allele = 218 base pairs
females (X,X) = homozygous
males (X,Y) = heterozygous
do viruses bug prokaryotes? do prokaryotes do or do not have viral DNA invading their genome? why? what about humans?
viruses do bug prokaryotes but they only have one circular chromosome
if the circular chromosome gets doubled by the virus, then it is not survival and natural selection will wipe the bacterium out
prokaryotes do have all the viral DNA invading their genome but do not have the space in the genome and cell to have all of the viral DNA because about 90% of their genome is to make proteins
humans have space in our chromosomes because we have 99% garbage in our chromosomes
can new gen sequencing be used for paternity testing?
yes but it is more expensive
what is inclusion for paternity?
occurs when the allele found in a child but not the mother; present in suspected dad
harder
what is exclusion for paternity?
neither of dad’s alleles at one locus appear in the kid
easier
what is the paternity index?
it is the likelihood of paternity and is calculated for each locus present
common allele has a low paternity index (PI)
rare alleles have high paternity index (PI)
all the test alleles together make a combined paternity index (CPI)
why does each loci have its own unique paternity index rather than sharing the same value?
they do not have the same index because it depends on the rarity of the sequences
some number of repeats will be more rare than others
how would you calculate CPI? what would the statement be?
multiply the relationship index numbers together
statement = dad is x times more likely to be the father than some random guy
what is probability of paternity (POP) and how is it calculated?
it is whether an alleged man is child’s biological father
calculated using CPI/CPI + (1-PP)
PP is prior probability and is 0.5
what is likelihood ratio? what is a high likelihood ratio? what is a low likelihood ratio?
compares the probability that 2 genotypes came from the same or different people
high likelihood ratio = came from the same person
low likelihood ratio = came from different people
what is genetic concordance? give an example
all locus genotypes (alleles) from 2 sources are the same
has all of our or their potential genotypes from the two sources are the same
ex. have four loci and they’re all the same then they have genetic concordance
what is the exception for paternity? why can’t it be applied to forensics?
mutational events or recombination events may generate a new allele in the offspring so one difference may not rule out paternity so further tests must be done
cannot be applied to forensics because an individual would have to mutate ALL their DNA after leaving their blood at the crime scene
what is match probability?
more loci analyzed, higher the probability, based on validation studies
what is a stutter? what does it look like?
PCR anomaly, polymerase missed a repeat because it gets confused, resulting in a different species in the amplified product
is a shorter peak compared to the other peaks
what is binning?
collection of all peaks or bands within a characteristic distribution of positions/one particular loci
what is host vs. graft when testing bone marrow engraftment?
host tissue attacks donor tissue
how are allogenic bone marrow transplants monitored?
using STR
what is an autologous transplant?
takes hosts own cells/own bone marrow and manipulate it in a culture (drug it) to improve the health of the tissue that is damaged and put the programmed cells back into themselves but is hard to program and culture the host cells
what is allogeneic transplant?
healthy donor cells that is transplanted into an unhealthy patient and can happen in organ transplants too
what is a recipient with donor marrow? elaborate
chimera which is cells with mixed genotypes which has some marrow that is the hosts and some of the marrow from the donor
chimera means to have different populations of cells adjacent to each other
why would you want to test chimeras after transplants?
to see how much of the patients cells are present and how much of the donor cells are present
what is the first part to chimerism testing? what are you looking for?
1) pretransplant informative analysis which is to find unique alleles/unique STR locations between the patient and the donor
NOT trying to see if they are a match
look for markers that could cause an immune reaction
the surface of the antigen on bone marrow cells which is what the immune system attacks
what is the second part of chimerism testing? what would be a graft failure?
2) post-transplant engraftment analysis
use STRs that were chosen to figure out what percent is the chimera
ex. donor and recipient peaks are in different locations before the transplant and after the transplant the peaks do not overlap
graft failure = is only the recipients peaks
how are polymorphisms used to measure engraftment after allogeneic bone marrow transplants? (written response question)
Polymorphisms, specifically STRs, are used to measure engraftment after allogeneic bone marrow transplants by figuring out what percent is the chimera from the chosen STRs. The donor and recipient STR peaks are compared with each other before the transplant and then are compared after to indicate whether there is a graft failure which is purely recipient STR or not.
what are informative loci and what would they be?
informative loci = donor alleles differ from recipient alleles
with the most informative loci, recipient bands or peaks do not overlap in donor bands or peaks
in desperate measures, if a recipient or donor band/peak matches one band/peak but does not have another band/peak then you can use it
what can a stutter be in a PCR reaction be and what would be the product of it? give an example
stutter can be a technical artifact of the PCR reaction in which a minor product of n-1 repeat units is produced
ex. 10 repeats but looks like 9 (1 less nucleotide = n-1)
what is noninformative loci?
no unique peaks but doesn’t mean it isn’t a good transplant match
it means that in order to analyze the success of the graft it wouldn’t be good methodology
would look at another donor to see if they’re a better match and need to look at how much host cells compared to donor cells
using informative loci, how are peak areas determined? why?
in florescence units or from densitometry scans of gel bands
in order test the success of the transplants, determine the areas under the peaks
accurate to detect what percentage of grafts or transplants are recipient vs donor
what is the formula for percentage of recipient DNA?
A(R) which is the combined area of the peaks/two peaks divided by A(R) + A(D) which is all four peaks x 100
what is the formula for percentage of donor DNA?
A(D) which is the combined area of the donor peaks divided by A(R) + A(D) which is all four peaks x 100
what is engrafted with different kinetics? why?
cell subsets (T, cells, granulocytes, NK cells, etc.) engraft with different kinetics
if looking at graft sets, need to look at a population of the hosts white blood cells
what was the article related to allogeneic bone marrow and G-CSF-mobilized stem cell transplantation?
it was able to predict the survival of a patient in allogeneic transplants by looking at granulocytes/particular population of granulocytes that came from the donor
what does analysis of the cellular subsets provide?
provides a more detailed description of the engrafting cell population and also increases the sensitivity of the engraftment assay
how are T cells used for predictive measures of transplant success?
on day 28, if patients have less than 90% of donor t-cells they have higher risks of graft rejection and disease relapse than those with more than 90% donor t cells
too many host t cells means that their immune system will attack the transplant/graft
what are the markers for chimerism analysis of cellular subsets?
T cells (CD3)
NK cells (CD56)
granulocytes
myeloid cells (CD13/CD33)
B cells (CD19)
stem cells (CD34)
what are the three methods for tracking transplant success? explain them
flow and immunomagnetic cytometric success (HLA typing) = pulling out cells magnetically and analyzing their STRs
immunohistochemistry and XY FISH (karyotyping)
DNA typing of polymorphisms = the best because unique polymorphisms are easier and faster to detect
what is split chimerism? what does it mean?
detection of different levels of engraftment in cellular subsets
means one cell subset looks good but another doesnt
what is linkage analysis?
gene-hunting technique that traces disease patterns in high-risk families
attempts to locate a disease-causing gene by identifying genetic markers that are co-inherited with the trait of interest like it being overexpressed or overrepresented
is usually specific to one area and can be performed in unrelated people (GWAS)
easier to scan for STR than for a point mutation in a large gene
what is linkage equilibrium?
two or more alleles that occur randomly in a population
alleles at different loci are inherited independently of each other
what is linkage disequilibrium?
STR never separated from phenotype and is not random
has the same weird mutation in a population/group that doesn’t exist much in society
found that there are some disorders where the number of STR repeats seem to be consistently the same
what is the one disease gene linked to STRs?
some of types of alcohol dependence is genetic
3 STR locations are overamplified a similar amount of times within a family, they found it linked to alcohol dependence
can test children if they have the alcohol dependence gene but only works if parent or one parent and a grandparent on the same side has had the same issue
what are Y-STR haplotypes and how are they inherited? (written response question)
Y-STR haplotypes are Y chromosomes with a set of markers, 23, that are inherited without any recombination. They can be used in various situations, such as sexual assault cases, parental lineage analysis, and identifying missing persons. Y-STR haplotypes are inherited paternally due to the Y-chromosome having no recombination during meiosis.
how are Y-STR haplotypes using in sexual assault applications?
the evidence collected from a crime scene usually has low amounts of male DNA in a high background of female DNA so had to create some particular Y profiles of kits that look at 23 good STRs
uses a multiplex assay with 6 different dyes called the Yfiler Plus kit that allows for amplification from multiple male-specific sample types
Yfiler Plus kit was designed for forensic labs to process highly challenging Y-STR caseworks like male-female mixtures
how are Y-STR haplotypes used for criminal ID? what does it depend on and how does it provide more evidence?
22 loci on the Y chromosomes raises discrimination index and has a common kit with 17 multiplex PCR primer sets
depends on controls like running relatives as well because the suspect and relatives will be genetically similar
if relatives are ran and they are not a match, it is a sign that it is not just randomness which provides more evidence to convict the suspect
define SNP, the technique to distinguish SNP, and the situations where you might want to detect SNPs (written response question)
SNP stands for single nucleotide polymorphism, which is a single-nucleotide difference in a DNA sequence. SNPs occur every 1331 bps in human DNAs, and 99% of them have no biological effect. SNPs occur in the expressed part of DNA, euchromatin, but not at a uniform density. The techniques that can be used to distinguish SNPs are sequencing, melt curve analysis, or allele discrimination probes. The situations where SNPs might want to be detected is identifying a human, mapping genes, gaining info about disease prediction, or analyzing chimerisms.
how many SNPs are within genes?
60,000 SNPs are within genes and some are genetic markers for disorders but a lot of the SNPs can be used to distinguish a sample of DNA
what are SNP haplotypes and how many bps contain how many SNPs?
SNP haplotypes are SNPs inherited together meaning they do not get unlinked from each other during recombination
20,000-60,000 bp undergo zero recombination contain about 60 SNPs
what are other applications of SNP analysis? explain them
human genetic variation as some studies have focused on SNPs related to this
HapMap which was aimed to identify SNP haplotypes throughout the human genome by trying to sequence a bunch of different ethnicities and creating maps of where SNPs are
up to 60,000 bp can be labeled with 4-5 tag SNPs which can provide information in high linkage disequilibrium areas
how is the HapMap used? what is it often used for and why? give an example
used to find reliable, taggable SNPs meaning a SNP will be present and always be present in that ethnicity
often used for medication failures and was first used for antibiotic resistance by figuring out what genes were mutated in antibiotic resistance
ex. would have a population where penicillin doesn’t work and a population where it does work and then look at SNPS where a particular location is different in success and failure
what is the process of using the HapMap when all the data is done?
download the data from HapMap and play with it
look for associations in the data from your family’s affliction to HapMap at selected SNPs
genetic association —> SNPs are genotypes in a population looking for trends
what is the HapMap linkage, disequilibrium plot?
confirms our DNA is ~99.5% ID across ethnic groups and allows to see genes or SNPs that are connected
easy analysis of data between ethnic groups
the red areas in the plot are unlikely to separate during recombination to create gametes
recombination seems to be different across different ethnic groups and the number of times
what are INDELs?
insertions or deletions of 1-10,000 bp
multiples of 3 are particularly uncommon in coding regions but relatively common in non-coding regions meaning natural selection has selected against INDELs being in coding regions
~200 - 230 frameshifting INDELS on each person and is between 16% and 25% of all sequences
INDEL frequency is markedly lower than that of SNPs except near highly repetitive homopolymers and microsatellites
what are LINES, SINES? are they frequently polymorphic? (written response question)
LINES are long interspersed elements that are retrotransposons that integrated into 20% of our genome. They are 6-8 Kbp long with a promoter and open reading frames that create a coding sequence for reverse transcriptase which is needed to create cDNA and integrate it into our genome. It creates its own enzyme to cut itself out and move in our genome.
SINES are short interspersed nuclear elements that are 100-700 base pairs long and are also retrotransposons but require LINE proteins to transpose. It is transcribed by RNA polymerase 3 and has internal promoters. They do not have enough sequence to have any open reading frames meaning they cannot make proteins for themselves so they steal polymerase info from other LINEs. SINES create genome instability or cancer as they are genomic parasites.
LINES and SINES are frequently polymorphic since they are retrotransposons. LINES can cut themselves out and insert themselves randomly into our genome and SINES have Alu elements that can jump around and cause disease in about 1 million copies of human genome; however, they often do not do anything as they can only jump to certain areas.
what are ERVs and how can they also cause polymorphisms? what have HERV genes been?
ERVs are endogenous retrovirus
majority of ERVs in vertebrate genomes are ancient, inactivated by mutation, and have reached genetic fixation in their host species and are extremely unlikely to have negative effects on their hosts except in unusual circumstances
mostly in the heterochromatin region
HERV genes have been coopted over 100,000 years of evolution for essential functions in immunity and pregnancy
what are mitochondrial DNA polymorphisms? what is heteroplasmy?
no recombination and changes only through mutations and is maternally inherited
has a faster mutation rate than nDNA which can result in heteroplasmy
heteroplasmy is when two or more mtDNA variants exist in the same cell making analysis challenging
it is circular DNA which gives it stability which may help in challenging specimens
mtDNA might be a whole new parasea with energy deficit disorders
where do the mutations tend to be in mitochondria? what is the rest of the genome and what do they code for?
mutations tend to be in the hypervariable regions 1 and 2
the hypervariable regions are in the D-loop which is an area that does not code for proteins
the rest of the genome are genes that code for 22 tRNA, 2 rRNA, and 12 genes for oxidative phosphorylation that makes ATP
what happens if oxidative phosphorylation gets mutated?
changes an individuals ability to make energy
will all relatives have the same mitochondrial sequences?
all maternal relatives will have the same mitochondrial sequences
what unique role does analysis of mtDNA play compared to nDNA? (written response question)
Since mitochondrial DNA is maternally inherited unlike nuclear DNA where it is inherited from both parents, mitochondrial DNA can be used to analyze the maternal lineage of an individual. In the hypervariable regions, sisters and brothers should be 99% matched. Mitochondria DNA can be typed as well for legal exclusion of individuals by looking at the hypervariable regions which are areas that are routinely sequenced in forensic analysis.
how was life originated on Africa confirmed by mitochondrial DNA? what does selection constrained regional mtDNA variation mean?
can trace the mitochondrial sequences and see where the cultures left and migrated
means that where cultures are isolated, their mtDNA stayed relatively similar to each other for the gene sequences and mtDNA would change depending on where the culture is isolated
what are other alternatives to DNA for polymorphic analysis?
protein identification as polymorphisms can exist here
epigenetic profiles by looking at methylation status
it is environmental and not always inherited
evaluated by methylation-specific enzymes and bisulfite sequencing
is there a biological exception to a positive ID by STRs?
identical twins