M5 Van- Desire for DNA - replication and transcription
1/8
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
9 Terms
why do we need DNA?
RNA mutates spontaneously- the deamination of cytosine into uracil is very common, and hard to detect because it is single-stranded and because U is one of the bases anyway (unlike in DNA)
it also has the 2’OH group, which allows it to form more H bonds and fold
RNA itself is generally stable, but is unstable in the current protein world because of the presence of RNAases
we need long stable sequences of information to produce enough proteins
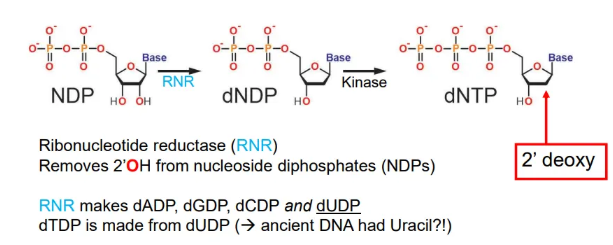
how are the DNA building blocks produced?
nucleoside diphosphates (NDPs) are converted into deoxynucleoside triphosphates (dNTPs) by ribonucleotide reductase (RNR), which removes the 2’OH, and kinase enzymes which add a third phosphate group
how does DNA replication occur?
DNA helicase separates the two strands by breaking the H bonds
complementary RNA primers are attached by primase at the start of the leading strand (replicated 5’ → 3’) and at regular intervals in the lagging strand (3’ → 5’)
DNA polymerase adds on the dNTPs following base pairing rules (condensation reaction releasing pyrophosphate), producing Okazaki fragments in the lagging strand
RNAse H degrades the RNA primers, and the fragments are extended until ligase joins the phosphate backbones together
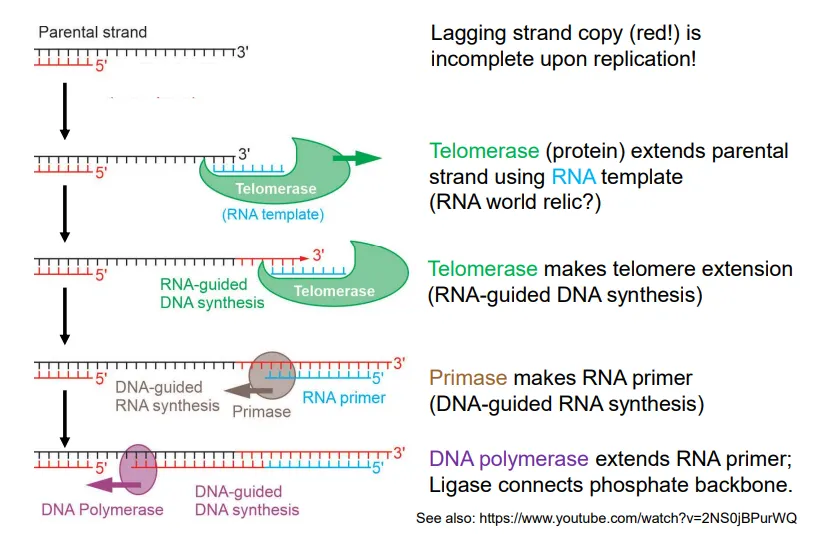
what happens at the end of DNA replication in the lagging strand in eukaryotes?
to form the final Okazaki fragment, telomerase extends the parental strand using an RNA template
primase attaches an RNA primer to this extended DNA strand
DNA polymerase extends the primer until the strands can be connected by DNA ligase
how is transcription initiated in prokaryotes?
the sigma factor (a cofactor of RNA polymerase) recognises and binds to the Pribnow and TATA box motifs in the promoter region upstream of the initiation site
this recruits RNA polymerase to bind to the DNA, produce a transcription bubble and begin RNA synthesis in the 5’ to 3’ direction using NTPs
the sigma factor dissociates

how is transcription terminated in prokaryotes?
after the stop codon is transcribed, termination signals found in the 3’UTR of the mRNA strand are also transcribed
these can either be:
inverted repeats, which cause hairpin loops (through base pairing) that will terminate transcription by RNA polymerase
a rut termination sequence, which is recognised by the rho protein that binds to RNA polymerase, terminating transcription

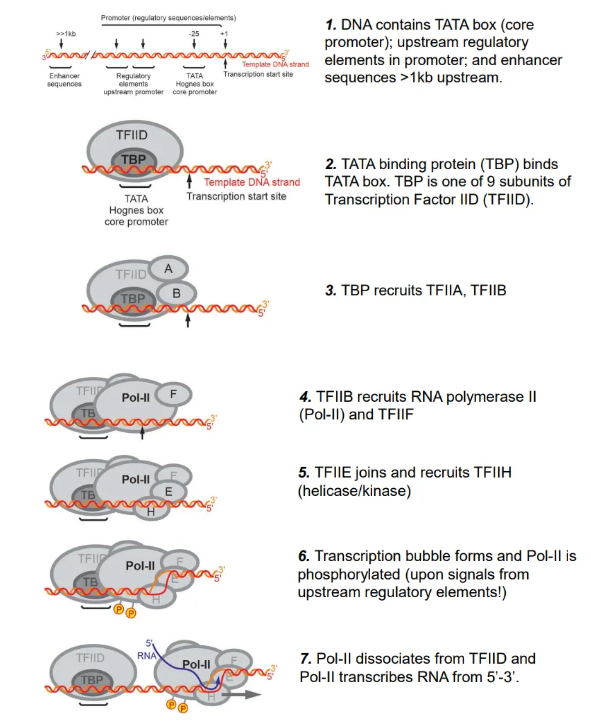
how is transcription initiated in eukaryotes?
the TATA binding protein (TBP), a subunit of transcription factor IID (TFIID), binds to the TATA box in the promoter region upstream of the initiation site
this recruits multiple proteins, including RNA polymerase II (Pol-II), which forms a transcription bubble, detaches from TFIID and begins to transcribe the RNA upon phosphorylation using NTPs
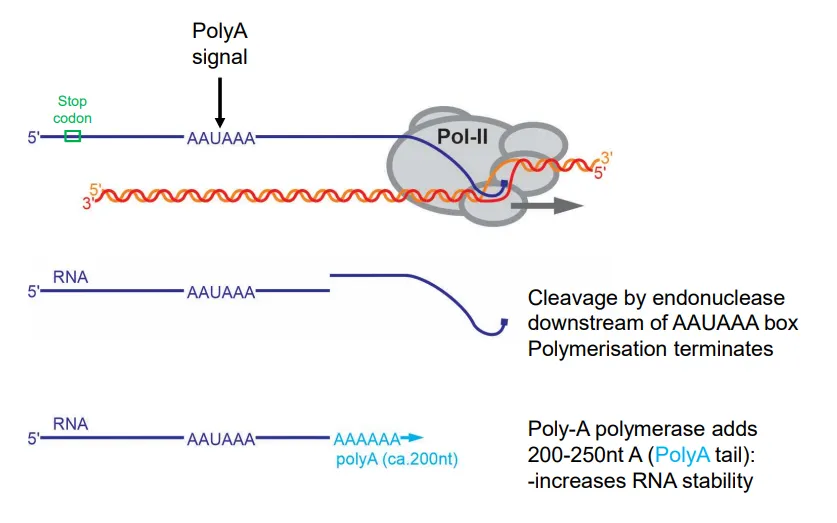
how is transcription terminated in eukaryotes and how is the mRNA modified?
after the stop codon is transcribed, a polyA signal (AAAUAAA) is found in the 3’UTR
this causes cleavage downstream by endonuclease enzymes, terminating transcription
a polyA tail (200-250 A nucleotides) is added to the 3’ end to increase the mRNA stability
the introns are then removed from this preRNA by splicing, catalysed by spliceosome (a ribozyme)
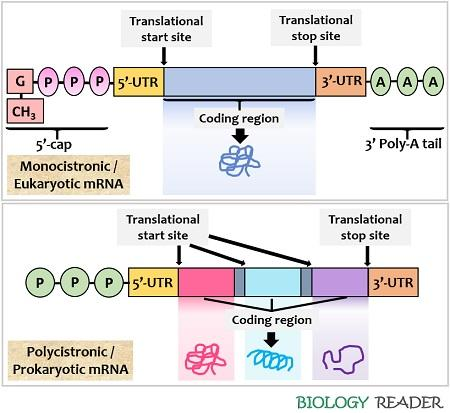
how is mRNA modified in prokaryotes and eukaryotes and why?
prokaryotes don’t have membrane-bound nuclei, so transcription and translation can occur simultaneously
at the 5’ end, they just have a triphosphate purine nucleotide
eukaryotic mRNA has to be transported out of the nucleus, so it is modified in a more complicated way:
a 5’ cap is added (made from guanosine triphosphate, and involving the methylation of the first two bases)
at the 3’ end a polyA tail is added (200-250 A nucleotides)
introns are also removed by splicing