Lab 2 Plasmid Miniprep Purification and PCR
1/28
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
29 Terms
fibroin-like protein
structural protein found in fiber
-------------------------------
In Lab (2) its: dragline silk of black widow spiders
how to express a synthetic gene
two experiments must be performed
1. use polymerase chain reaction (PCR) to amplify the synthetic gene sequence
2. prepare the prokaryotic expression vector for the insertion of the PCR product
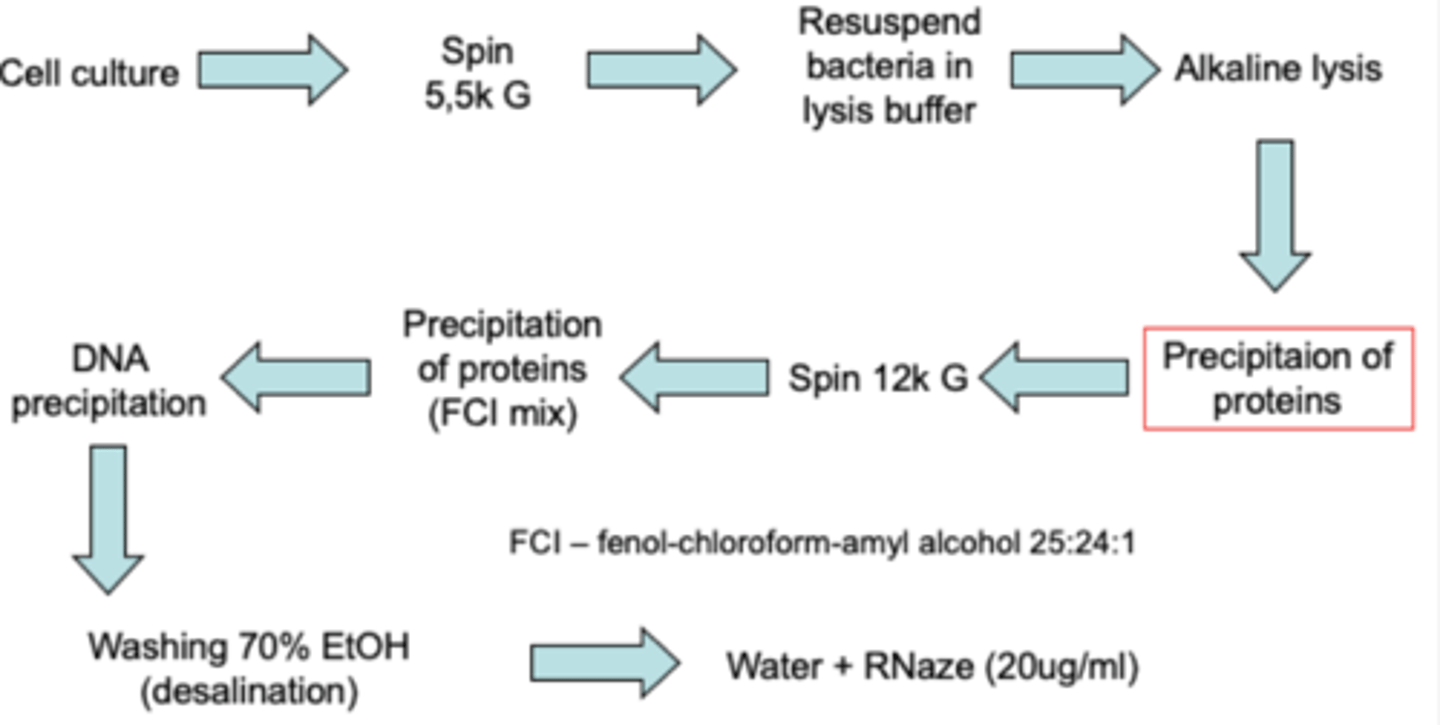
Plasmid DNA isolation steps
FROM START TO FINISH
1. start with liquid culture
2. break open though an alkaline lysis step
3. separate plasmid DNA from proteins and host DNA by neutralization
4. bind plasmid DNA to a silica resin and wash the plasmid DNA
5. elute clean plasmid DNA
preparation of Bacterial Lysate
1. Start with a liquid culture of bacterial cells (filled with LB/nutrients and ampicillin)
-------------------------------
2. concentrate bacterial cells by centrifugation in an Eppendorf tube (done twice/Double pelleting) to ensure maximum plasmid DNA yield
-------------------------------
3. Result: pallet at the bottom of tube
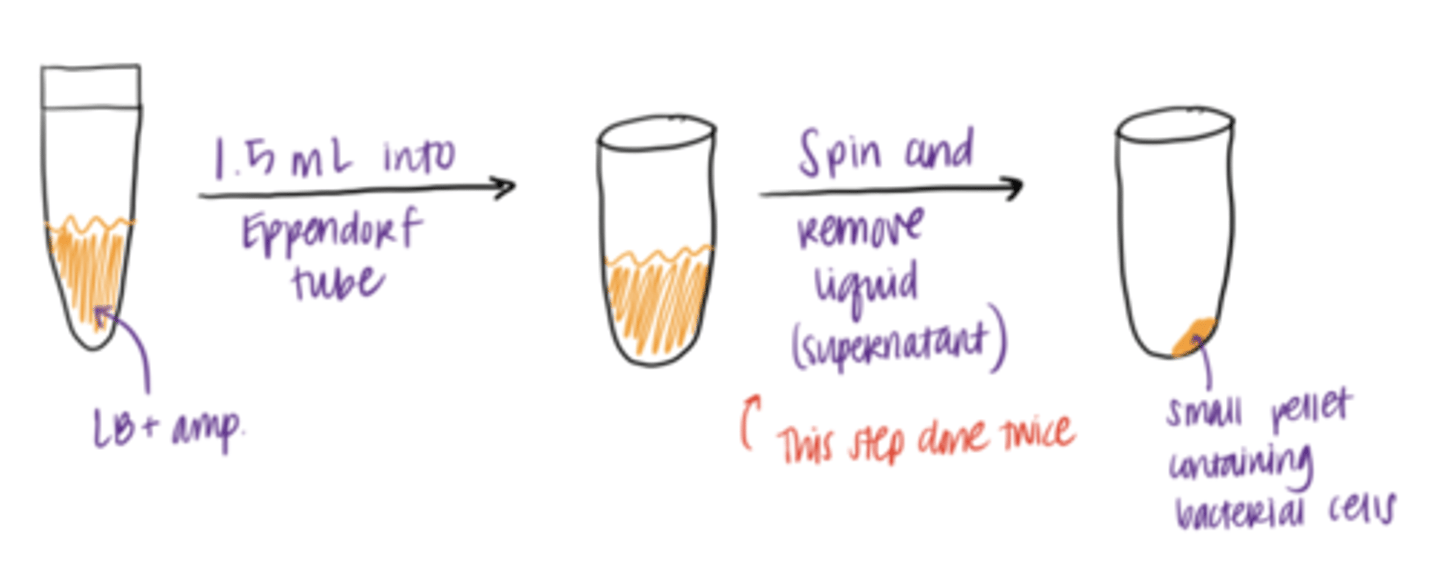
Why is it good for cells to grow overnight?
it ensures the cells contain large amounts of plasmid DNA in their cytosol

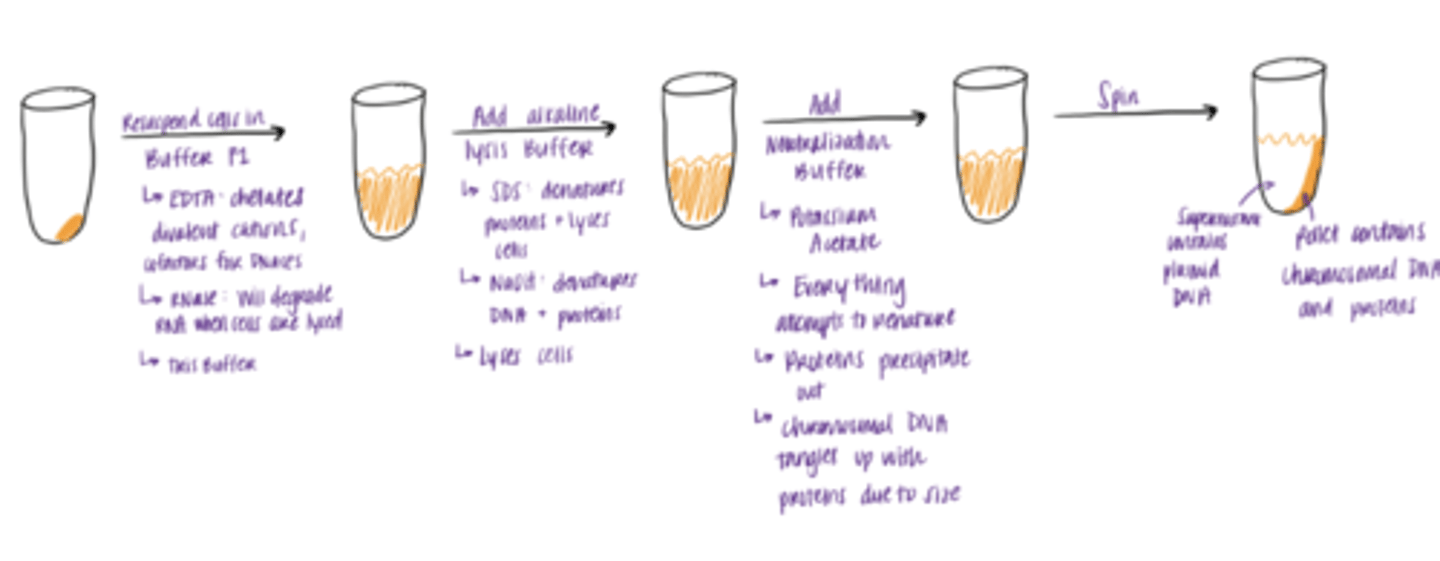
Clearing of the bacterial lysate
1. Resuspend pallet in Buffer P1
-------------------------------
2. Lyse cells by adding Buffer P2
-------------------------------
3. Neutralize solution by adding Buffer N3
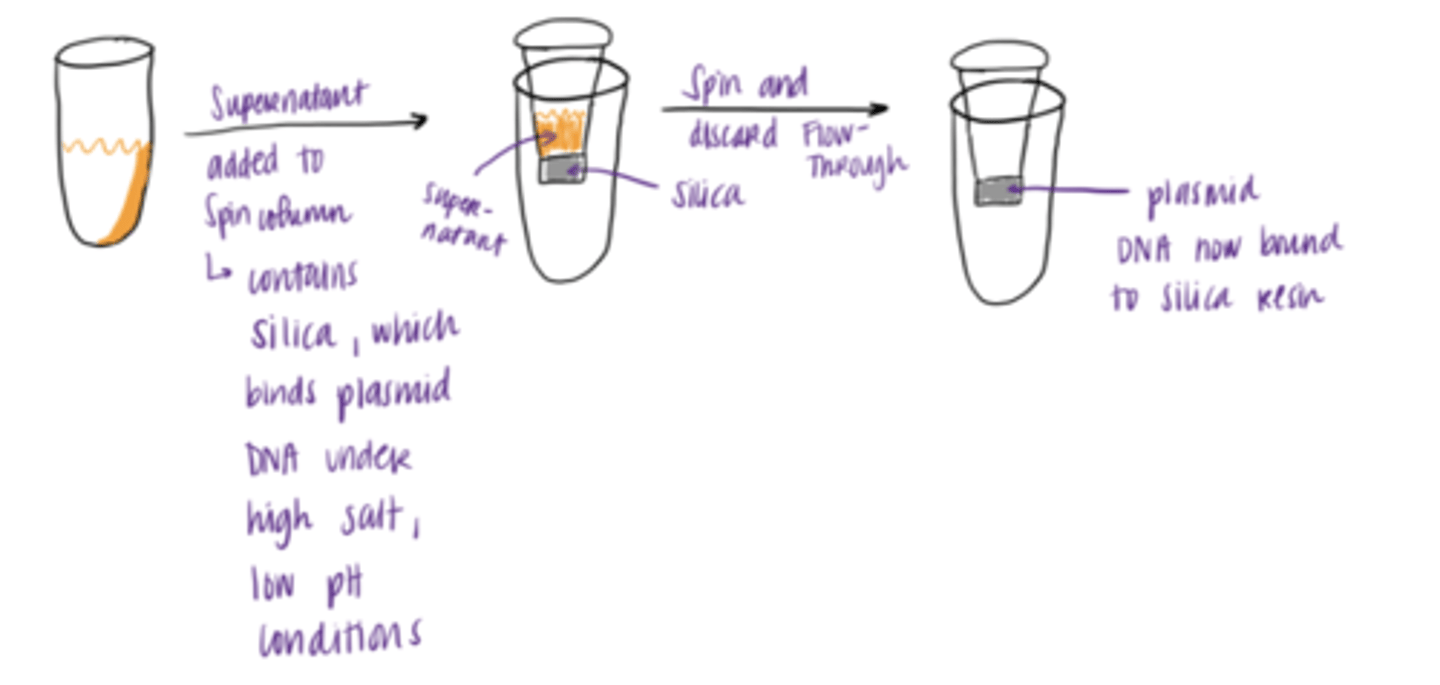
absorption of DNA onto silica
1. supernatant is added to spin column
-------------------------------
2. spin and discard flow through
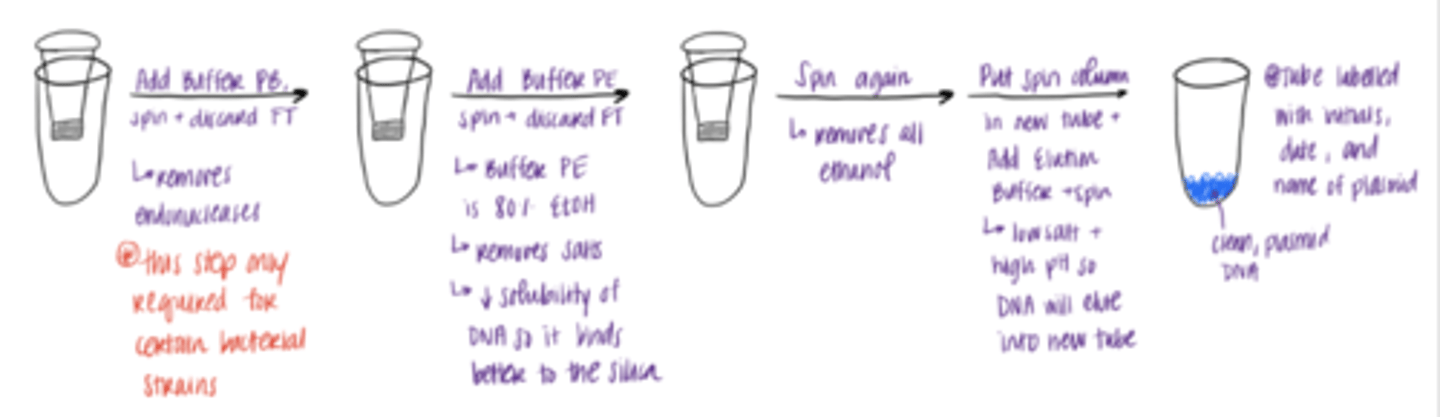
washing and elution of plasmid DNA
1. add Buffer PB to spin column, spin, and discard flow through (FT)
-------------------------------
2. Add Buffer PE, spin, and discard FT
-------------------------------
3. Dry spin (centrifuging without adding anything to the column to remove all the ethanol)
-------------------------------
4. transfer spin column to empty tube, add Buffer EB, and spin
-------------------------------
RESULT: PURE PLASMID DNA!
plasmid DNA
is normally found as double stranded DNA molecules that replicate when bacterial cells divide by the process of binary fission
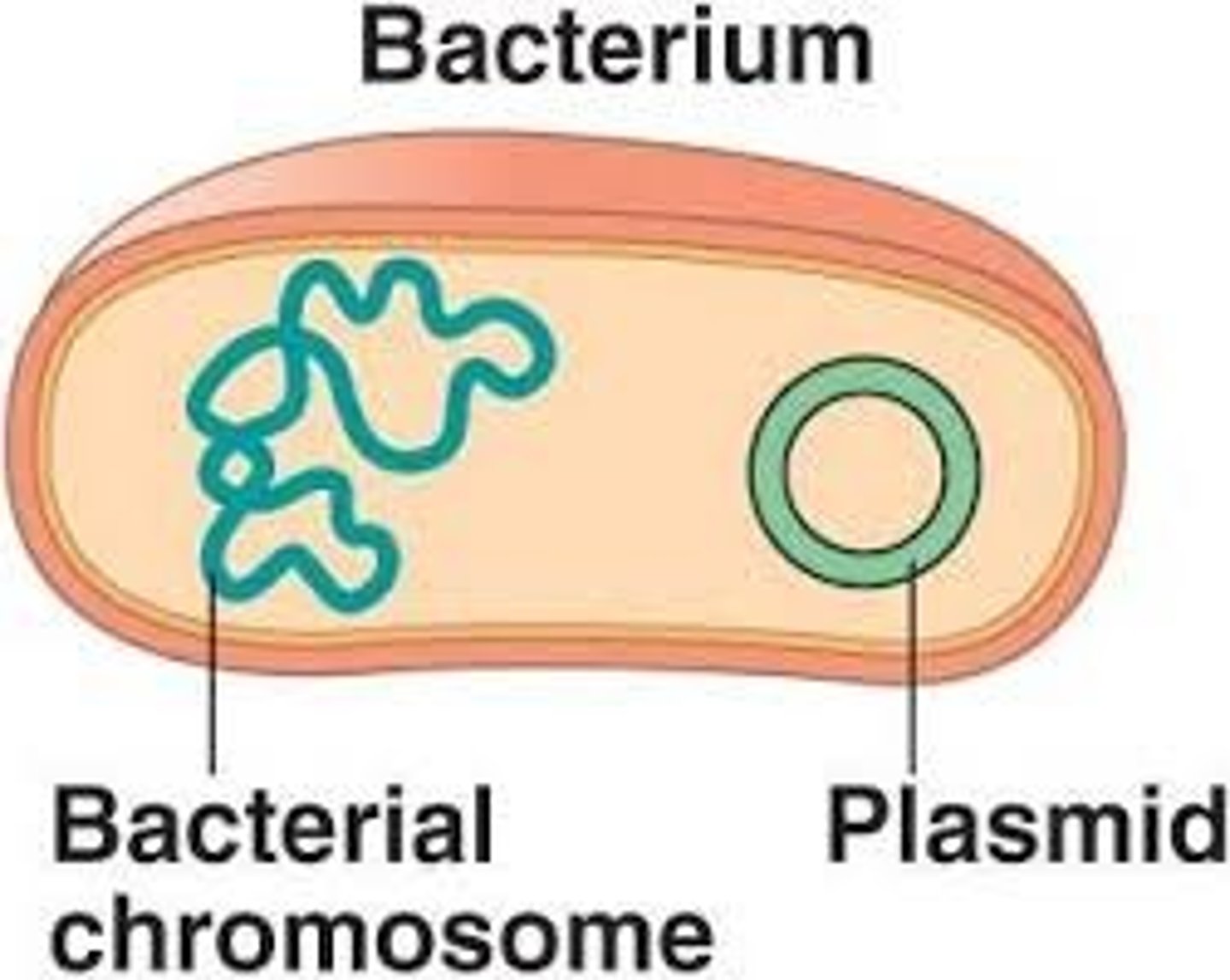
How can you retrieve the plasmid DNA sample?
1. break open the cells and use a purification method to separate the cloning vector from the host genome, RNA, protein and other bacterial macromolecules
alkaline lysis
uses SDS to break up phospholipid bilayer and NaOH to dissolve structural proteins
-------------------------------
HANDOUT notes:
- takes advantage of the fact that the plasmids are relatively small, supercoiled DNA molecules and bacterial chromosomal DNA is much larger and less supercoiled
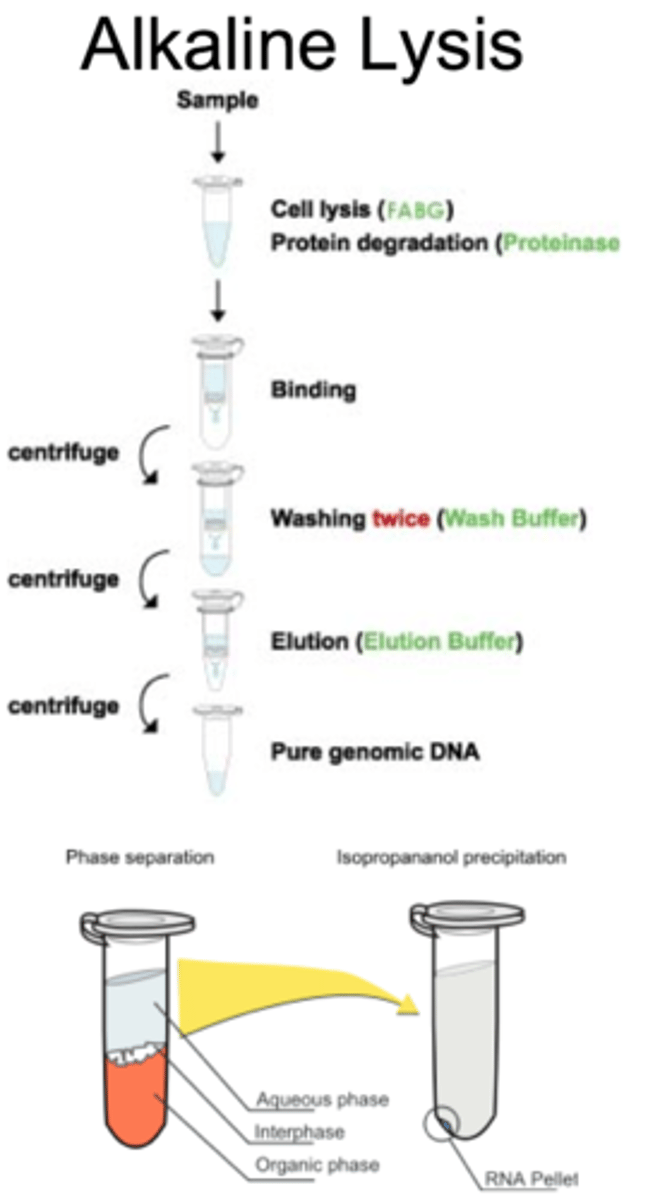
alkaline lysis procedures
Based on handout:
1. preparation and clearing of the bacterial lysate
2. absorption of DNA onto a QIAprep membrane
3. Washing and elution of plasmid DNA
why are bacterial cell lysed under alkaline conditions?
to denature both nucleic acids and allow the chromosomal DNA and proteins to precipitate and renature correctly
double pelleting
pelleting twice
-------------------------------
WHY IS THIS DONE?
- to increase your overall plasmid DNA yield at the end of the procedure
- this procedure collects the bacterial cells which are suspended in the liquid medium into a cell pellet at the bottom of your tube
What happens to the cells when they are re-suspended in a buffered solution with RNase and are lysed?
the RNase will catalyze hydrolysis of all the RNA molecules into nucleotides, but the DNA will not be affected
SDS (sodium dodecyl sulfate)
ionic detergent which disrupts cell membranes and destabilizes all hydrophobic interactions holding various macromolecules in their native conformation
Buffers used in Lab 2
- Buffer P1
- Buffer P2
- Buffer P3
- Buffer N3
- Buffer PB
- Buffer PE
- Buffer EB
Buffer P1
Resuspension Buffer
-------------------------------
FEARTURES/CONTAINS:
- RNase A: (breaks down RNA
- Tris-HCL: (common buffer to keep solution at the right level)
- EDTA: chelate divalent cations (when removed it stopes DNase from degrading the DNA)
- Lysosome: digest the bacterial cell wall
Buffer P2
alkaline lysis buffer
-------------------------------
FEATURES/CONTAINS
- SDS: ionic detergent (soap)
- function is to lyse the cell and denature the proteins
- NaOH: used to denature the proteins
Buffer N3
Neutralization
-------------------------------
FEATURES/CONTAINS
-KAcO pH of 5.5: (3 M helps precipitate the DNA and denature the chromosomal DNA)
Buffer PB
remove endonucleases so that the plasma doesn't get destroyed
Buffer PE
remove salts
-------------------------------
FEATURES/CONTAINS
- 80% ethanol (decreases the DNA solubility and increases the DNA binding)
Buffer EB
High pH and Low
-------------------------------
FEATURES/CONTAINS
- elutes the DNA from the column so it is ready for future applications
PCR (polymerase chain reaction)
extremely powerful technique that can amplify billions of copies of DNA starting from very little material
-------------------------------
WHAT IS IT USED FOR IN BIOLOGY?
- amplify genes for cloning pirposes
- introduce mutations into genes
- amplify nucleic acids for DNA sequencing
- modify ends of gene sequences to facilitate insertion into cloning vectors
What is PCR used for in biology?
- amplify genes for cloning purposes
- introduce mutations into genes
- amplify nucleic acids for DNA sequencing
- modify ends of gene sequences to facilitate insertion into cloning vectors
TmS
is defined as the temperature at which 50 % of primers are docked to the template
what happens if the annealing temperature drops too low below the primer's Tm
the primers can "mis-prime" and lead to spurious amplification of the wrong piece of DNA sample
Taq polymerase
lacks a proofreading function and has an unusual property of adding a non-template derived nucleotide to the 3' ends of the dsDNA molecule
Q5 High Fidelity DNA Polymerase
has proofreading activity and lacks the intrinsic ability to add a non template derived nucleotide to the 3' ends leaving them completely blunt ended