Gene Transfer
1/77
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
78 Terms
Whats horizontal gene transfer?
Movement of genes between cells that aren’t direct descendants of another as opposed to vertical transmission (mother to daughter cell)
Allows cell to quickly acquire new characteristics and fuels metabolic diversity → aids in evolution and adaptability of a bacterium
Useful molecular tools in bacterial genetics
What are the 3 mechanisms of genetic exchange known in bacteria?
Transformation
Transduction
Conjugation
How many directions does DNA transfer occur in?
Typically occurs in one direction from donor to recipient
Whats transformation?
Free DNA released from one cell is taken up by another
There are 2 versions of transformation
Natural
Artificial / chemical
Whats transduction?
DNA transfer mediated by a virus/ a phage
Bacterial DNA is mistakenly packaged into a phage capsid (instead of viral DNA)
Defective phage then released to infect another cell injecting bacterial DNA
Whats conjugation?
DNA transfer requires cell-to-cell contact and a conjugation plasmid in the donor cell
Donor DNA is transferred through a piles
Who did the discovery of transformation and why was it important?
Griffith did in 1928 with his experiment using Streptococcus pneumoniae
Certain strains produced a capsule
It was important because it provided key evidence that DNA was genetic material
What’re capsules and explain their qualities?
Capsule is composed of sugar units linked to form a polysaccharide material
Different forms of S. Pneumoniae affect virulence (I.e. ability to cause disease)
Capsule allows bacteria to evade attack by immune system
Bacteria gets destroyed by immune system if no capsule
Chemical composition of capsules can vary slightly between different smooth strains of S. Pneumoniae
Ex: type II different from type III depending on type of sugar in capsule
Can have reversion between smooth (S) and rough (R) but only within a type
Ex: type IIS ←> type IIR but not type IIS ←> IIIS
Explain what occurred in Griffiths experiment
He used type IIR cells and type IS cells
R killed mice, S didn’t kill mice, R transformed to S killed mice
IIR cells were transformed into IS cells
Later experiments showed that:
Living S cells could be produced in liquid culture of a mixture of R and heat-killed S cells (I.e. no host required)
Transforming activity was retained in filtered extract of dead S cells (I.e small molecule)
What did Griffith hypothesize?
Griffith theorized that specific protein structure was responsible for transformation
What did Avery, McCloud and McCarty (1944) do?
Proved that DNA was the agent behind transforming activity
Reproduced Griffith’s experiment, cell extract was treated with protease, DNAse and RNAse
Proved that transforming principle was DNA
DNAse = doesn’t kill mouse, everything else kills mouse
What is natural transformation?
Capacity to take up free DNA is genetically determined (e.g. Vibrio, Neisseria, Bacillus, Streptococcus)
When a cell lyses, chromosomal DNA leak out of the cell and due to its large size, fragments (~10-15 kbp)
What does natural transformation require?
DNA from donor cells must be fragmented linear dsDNA
Does not work with ssDNA, only has to be linear dsDNA
Recipient cells must be competent and take up the dsDNA fragments
Should be able to make competence factors:
Excreted from cell - reaches effective concentration at high cell density
Binds to surface receptors - induces synthesis of other proteins involved in the transformation
Occurs during late log phase
Explain how competence is directly linked to pili?
Pili binds and facilitates uptake of DNA into cell by retraction
Mechanisms exist to account for differences in cell envelope structure for gram negative and gram positive bacteria
What’re the steps of transformation?
Transformation complex binds exogenous (donor) dsDNA
Uptake of DNA. As dsDNA is brought into the transformation complex, one strand is degraded by nucleases and ssDNA enters into the cell. Need dsDNA to function at all, as it enters the cell, only ssDNA enters cell
Homologous recombination
The ssDNA fragment will be paired with a homologous region of the chromosome
Pairing is mediated by RecA
Strand exchange will occur
Transformed cell
Donor DNA is integrated into chromosome of recipient cell
Successful recombination usually by a change in phenotype
Ex: donor DNA from Trp+ cells taken up by competent Trp- cells
dsDNA converted into ssDNA, recombination, change genotype and hence phenotype
Whats something that happens in natural transformation?
ssDNA is bound by SSB
RecA displaces SSB and promotes strand invasion of chromosomal DNA
Double cross-over event with a ssDNA fragment and chromosomal DNA
Recombination with heteroduplex region resolved by either DNA repair or replication
What happens in the homologous recombination model?
In E.coli, the RecBCD pathway will mediate homologous recombination between dsDNA linear fragment and chromosomal DNA
RecBCD is a complex that has endonuclease and helicase activity
Displaced ssDNA is coated with SSB
RecA will displace the SSB protein binding the ssDNA region
RecA will mediate strand invasion of the double stranded chromosomal DNA
The cross-strand complex is not static; complex will move until strand homology is found (region of 100s to 1000s of bases)
Cross-strand complex will then be cleaved on a horizontal or vertical plane - a random event done by a resolvable enzyme
What does resolution of cross strand complex result in?
Two products
Patches - no recombination (no exchange of markers) with short heteroduplex regions
Splices - recombination has occurred (exchange of markers) with short heteroduplex regions
Heteroduplex regions are mismatched regions which resolved by DNA repair or by DNA replication (2 molecules each with slightly different sequence)
Are all bacteria transformable?
No, E.coli isn’t naturally transformable
Does the process of transformation differ from groups of bacteria, if so how?
Yes
Some bacteria have specific sequences within the DNA to recognize “self” → protective mechanism against incorporation of foreign DNA into its chromosome compromising genetic integrity
Ex: Haemophilus influenza
Doesn’t require competence factor
Receptors on cell surface to recognize a specific 11-bop DNA sequence (occurs ~600 times within chromosome)
Only linear dsDNA with this sequence will be taken up by cell
What happens to bacteria that’s not naturally transformable?
They can be made transformable artificially by chemical treatment or exposure to electric current
Done to uptake plasmids (circular dsDNA)
Plasmids will not recombine with the chromosomes unless specifically designed to do so
Plasmids will replicate independently and be passed onto progeny cells
Linear dsDNA fragments introduced through the pores will be degraded by host defense systems that protect against incorporation of foreign DNA
Why is transduction known as a universal mode of gene transfer?
Because its theoretically possible for all bacteria as long as there’s a compatible phage
Whats a transducing particle?
Host-derived DNA carried from one cell to another inside a phage capsid
Phage that has picked up bacterial DNA (carry 1 -2 % of bacterial genome; 1 fragment is ~35-80 genes)
What are the 2 types of transduction?
Generalized transduction
Specialized transduction
What happens in generalized transduction?
Only bacterial DNA is carried
Any gene can be transferred from the donor cell
What happens in specialized transduction?
Some bacterial DNA and some phage DNA is carried
Only specific genes can be transferred from the donor
What happens when a transducing particle infects a recipient cell?
Only steps 1 and 2 are followed as this is mediated by capsid proteins
Lytic or lytic/ lysogenic cycles aren’t followed due to lack of phage genes
Whats a transducing phage?
Phages capable of transduction (not all phages can do this)
Whats a donor strain?
Original bacteria strain in which transducing particle had multiplied and picked up host bacterial DNA
Whats a transductant?
A cell that has been transduced (a.ka. Recipient strain)
What must a phage be to be a generalized temperate phage?
Can be temperate or lytic
Must not completely degrade host DNA because there will be no DNA to package into capsid if it degrades
T4 phage is not a good choice (obliterates host DNA)
Need to have not too specific pac or cos sites for packaging of DNA otherwise host DNA will not be packaged
Also broad host range of adsorption for possibility on wide range of bacterial species
What’re the 3 possible fates for injected DNA in generalized transduction?
Host restriction - degraded by host enzyme system (i.e restriction enzymes)
Abortive transduction - not degraded, but fails to recombine
Diluted out of population by successive cell divisions
Stable gene transfer - recombined into the chromosome by:
Pairing of exogenous dsDNA with homologous region in chromosome
Unlike transformation, both strands are replaced by homologous reciprocal recombination
What’re the steps of generalized transduction using E.coli?
Trp gene as marker, Donor strain is Trp+ and recipient strain is Trp- (can’t synthesize tryptophan so autotrophic for tryptophan)
Infect donor strain (Trp+) with P1 linear dsDNA phage under conditions that will promote lysogeny
Expose lysogeni culture to UV light to induce lytic cycle
P1 reproduces
Host cell lyse, releasing progeny and lysate which is just the debris present after lysis
Most are normal phages but some are transducing particles with ~1 in 10000000 carrying the Trp+ gene
Briefly expose lysate to UV light
Inactivates normal P1 phages by reducing its ability to reproduce but has little effect on the transducing particle
Mix phage lysate with recipient (Trp-) strain at MOI slightly less than 1
Each cell should be infected by no more than 1 phage or particle
Reduce the likelihood of a cell infected by both a transducing particle and a normal phage
Selecting transducants (cells which have been transduced)
Plate infection mixture on a selective medium → minimal medium without tryptophan
Trp- recipient cells fails to grow
Only cells that recieved Trp+ will grow (transducants)
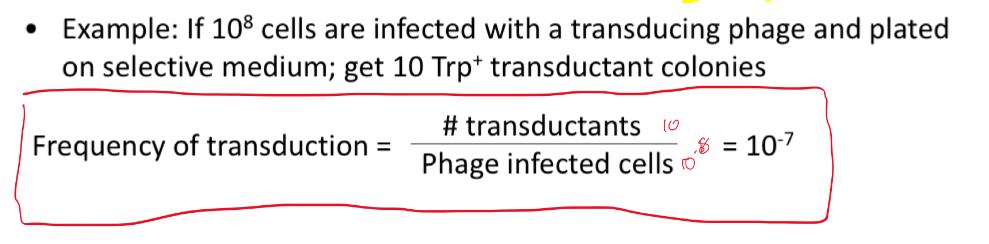
What is the frequency of transduction?
Ratio of successful transductants/ phage infected cells
Whats the frequency of transduction?
What causes pinpoint colonies on the same plate?
Abortive transduction
Ex: Trp+ gene has been transferred, but fails to recombine
When the merodiploid (2 copies of gene Trp+ and Trp-) divides only one cell receives Trp+
Extra chromosomal dsDNA fragment eventually degraded
What is co-transduction frequency?
Transfer of 2 markers on same fragment of transducing DNA
Frequency depends on distance between 2 genes
Closer together - more likely they will be co-transduced
Historical method of mapping genes
Whats an example of co-transduction frequency?
Donor is bio+ and gal- and recipient is bio- and gal+
Perform transduction experiment
Select only for bio+ transductants → 100 bio+ colonies are counted of which each may be Gal+ or Gal-
Bio is the selected marker, gal is unselected marker
What is specialized transduction?
Transfer of specific genes
Uses temperate phage that have specific insertion sites in the host chromosome
Only genes close to the prophage insertion sites are transduced
Due to aberrant (incorrect) excision of the prophage
In specialized transduction what happens when lysogenized E.coli is exposed to UV?
Expose lysogenized E.coli briefly to UV → induction → collect lysate
Phage progeny are mostly normal lambda phage
Specialized transducing particles arise at low frequency (~1 in 10^6 phages) due to aberrant excision
Recombination doesn’t occur at normal recombination sites
What does an excised prophage include?
Some bacterial genes, but excludes some phage genes; limited by phage head sizes
Cell still contains all phage genes - phage reproduction can complete
What is specialized transduction dependent on?
Dependent on which side cross-over happens
Gain gal genes but loses head/ tail genes
Can’t complete virion assembly
Follows lysogenic pathway
Or
Gain bio genes but loses xis and int
Can be assembled and released
Can’t become lysogenic
Whats a gene transfer agents?
They resemble tiny tailed bacteriophages and contain random small pieces of host DNA
They are NOT considered true bacteriophages as they don’t contain genes encoding their own production and do not produce characteristic viral plaques
Genes encoding GTA’s lie within the genome of the cell that produces them, while other regions of the genome are packed within the agents
Common amongst marine bacteria
Seems to have evolved as a mechanism for a subpopulation of cells to sacrifice themselves in order to dispose genes in a protected manner
What is conjugation?
Form of horizontal gene transfer that requires cell-to-cell contact
Plasmid-encoded conjugation mechanism that can mediate DNA transfer between unrelated cells, even between different genera
Transfer copies of themselves and the genes they encode (eg: antibiotic resistance) to new host cells
Conjugation plasmid
What does conjugation require?
Process requires a donor cell (containing the conjugative plasmid) and the recipient cell (doesn’t contain conjugative plasmid)
Transfer mechanisms may differ depending on the participating plasmid, but most plasmid in gram-negative bacteria employ similar mechanism
Conjugation has also been demonstrated in a few gram-positive bacteria (streptococcus)
When was conjugation discovered?
Initially discovered by Lederberg and Tatum (1946)
Mixed 2 different auxotrophic strains, incubated for several hours and plated on minimal medium
Growth of several prototrophic colonies
Genetic exchange occurred
Due to Hfr strains
What did Davis discover in 1950?
Determined that physical contact is necessary for genetic exchange
Separated 2 auxotrophic strains by a fine filter which caused genetic exchange to not take place
Which bacteria can conjugate and which ones can act as donors?
Many but not all bacterial species can conjugate
Only certain bacterial strains can act as donors
What is the F factor?
Conversion of non donor strains to donor strains
F- to F+
What does F plasmid encode?
It encodes a pious
F pious of the donor (F+) makes contact with specific receptors on cell surface of F-
Pious contracts bringing the cells together
Forms a conjugation bridge
What is the F plasmid?
Fertility plasmid, self replicating circular dsDNA
~100 kbp long
Primary function is to self transfer to F- cells
F- cells don’t have plasmid, F+ transfers plasmid to F- cells to make them all F+ cells, happens in ~60 min
What are the features of the F plasmid?
tRNA region: ~21 genes - encode proteins for F-pious and transfer of plasmid
OriV: origin of replication
Or IT: origin of transfer
Insertion sequence elements IS2 and IS3, and transposing Tn1000
~1kbp in size
Also present on bacterial chromosome
Whats the conjugation mechanism for the F pilus?
Pilus is assembled and establishes cell contact
One strand of the F plasmid is nicked at orbit
One strand becomes linear with 5’ and 3’ ends
The 5’ end is displaced and enters conjugation bridge
Transferred to F- as ssDNA
The 3’ end is extended using intact circular strand as template
Rolling circle replication
After 1 complete replication, linear strand is cut
Inside the recipient cell: ssDNA is replicated to make dsDNA
Replication proceeds 5’ to 3’
Okazaki fragments
dsDNA is formed
Double stranded ends joined to make circular F-plasmid
When does the Hfr strain occurs?
Occurs when F plasmid can integrate into the host chromosome, this is called an episome
Episome = plasmid that can integrate itself into host chromosome
IS2, IS3 and Tn1000 can act as integration sites
F+ cell is converted into Hfr (high frequency of recombination) cell
Can be isolated as pure culture
F plasmid can mediate transfer of chromosomal genes
F factor genes are expressed producing F pili therefore able to conjugate with F- cells
What happens when conjugation begins in Hfr strain?
Transfer begins at orbit
Located in the middle of the F factor
Only about half of the F factor is transferred
Followed by chromosomal genes
To transfer the 2nd half of the F factor, the entire chromosome of the donor would need to be transferred to recipient
How long does the complete transfer of Hfr strain take?
Takes ~100 minutes
Very rarely happens as conjugation bridge must remain in place
Thus, in an Hfr x F- cross, the recipient cell always remains F-
Whats the mechanism of transfer in Hfr strain?
One strand is nicked at oriT
oriT is at center of F plasmid
F factor genes constitute both the 3’ and 5’ ends
5’ end of the linear strand is transferred to the recipient
3’ end serves as primer for DNA synthesis to replace transferred strand
Transferred as ssDNA and them converted to dsDNA in recipient
Half of F factor
Followed by chromosomal genes
But conjugation bridge often breaks which results in a fragment of transferred DNA
To become a stable genetic element, it must be recombined into a chromosome
How were Hfr strains used to map the E.coli chromosome?
There are several possible insertion sites on E. Coli chromosome
Location of the F factor is different for each Hfr strain
Can be located on either strand
Transferred in either direction
Each Hfr strain can be used to map about 25-40% of the chromosome by controlling the breaking of the conjugation pilus at timed intervals
LOOK AT TIME MAPPING SLIDES
What happens when overlapping gene orders are placed together?
Overlapping gene orders can be placed together to form a composite map
Distance on map given in units of time (ie. min)
Total length of E. Colin’s chromosome is 100 min
If chromosome is 4500 kbp long (so each min = 45 kbp)
How many cells in a Hfr strain will be F+?
A very small number of cells in any Hfr strain will be F+ (complete transfer is rare), but excision of F (factor) plasmid can occur
Can F factor be excised incorrectly, if so how?
Yes
Some chromosomal genes are included on the plasmid
Some F plasmid genes are left behind
These cells are known as primary F’ donors
Can transfer specific genes at a very high frequency
How does gene transfer by F’ donors occur?
Primary F’ donors can form a pilus
All F factor genes are present
F’ x F- conjugation
Whats the result of transfer by F’ donors?
Plasmid (and any chromosomal genes on it) will be transferred at very high frequency
Recipient gains the entire plasmid and becomes a secondary F’ cell
Stable partial diploid or merodiploid (2 copies of a gene)
Can be used for complementation tests to see if strain with deletion has impact on phenotype
What are F’ factors containing bits of lac operon used for?
They were used to derive function of lac promoter, operator and genes
Make chromosomal mutations in lac operon, and complement function by conjugating F’ factor containing normal copies of lac genetic elements into mutant strains
Derived gene function by rescued phenotype
This was how the lac operon was studied in the 1950s and 1960s
Where is mobile DNA: transposable elements found?
Commonly found in chromosomes, plasmids, and viral genomes
What are mobile DNA: transposable elements?
Pieces of DNA that can move from place to place in the genome
Move by a process called transposition
Frequency ranges from 1 in 10³ to 1 in 10^7 pre element/ per cell generation
Important both in natural genome rearrangement and in genetic analysis
What are the qualities of mobile DNA: transposable elements?
Do not require homology with the destination site
Do not possess their own origin of replication
Replicated when the host DNA molecules into which they are inserted is replicated
What are the 2 types of transposable elements in bacteria?
Insertion sequences (IS/ transpons)
Composite transpons
Explain what Insertion sequences (IS/ transpons) are
Short DNA segments of ~1000 nucleotides long
Typically contain inverted repeats of 10-50 base pairs
Encode only 1 protein → transposable (helps it move from one place to another)
Explain what composite transposons are
2 IS elements flanking antibiotic genes of other genes providing beneficial attributes
Has additional genes encoded within between IS elements (eg: antibiotic resistance)
Often the transposable in one of the IS elements is dysfunctional to allow movement of the whole as a unit
Explain the movement of IS elements and composite transposons
Transposase recognizes inverted repeats and moves transposable elements from one site to another
What are the 2 mechanisms of movement of IS elements and composite transposons?
Conservative transposition
Replicative transposition
Explain conservative transposition
Example: Tn5
Transposase recognizes inverted repeats and cuts on either side
Transposon is removed from original DNA molecule
Carried to a target site 5-9 bp sequence randomly selected
Cuts the target DNA by making staggered nicks
Inserts the transposon
Staggered cut results in gaps on either side
Gaps are filled by DNA pol 1 and DNA ligase
Results in direct repeats flanking the transposon
Is direct repeats a part of the transposon?
NO, direct repeats is not a part of a transposon but a result of the transposition event
What is the idea behind replicative transposition?
Transposon that carries an additional gene encoding enzyme resolvable
Transposon is replicated as it jumps, so that after transposition, both donor and target DNA molecule have a copy of the transposon
What are the steps of replicative transposition?
Example in Tn3
Transposase cuts staggered nicks on either side of transposon
Also cuts staggered nicks on either side of target site
Strands are exchanged and lighted
2 DNA molecules joined together With gaps corresponding to the target site and the transposon
The gaps are filled in by DNA polymerase and sealed ligase
Resolvable catalyzes recombination
Resolved into 2 separate DNA molecules, each with a copy of the transposon