Kaplan MCAT Biochemistry Chapter 7: RNA and Genetic Code
1/48
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
49 Terms
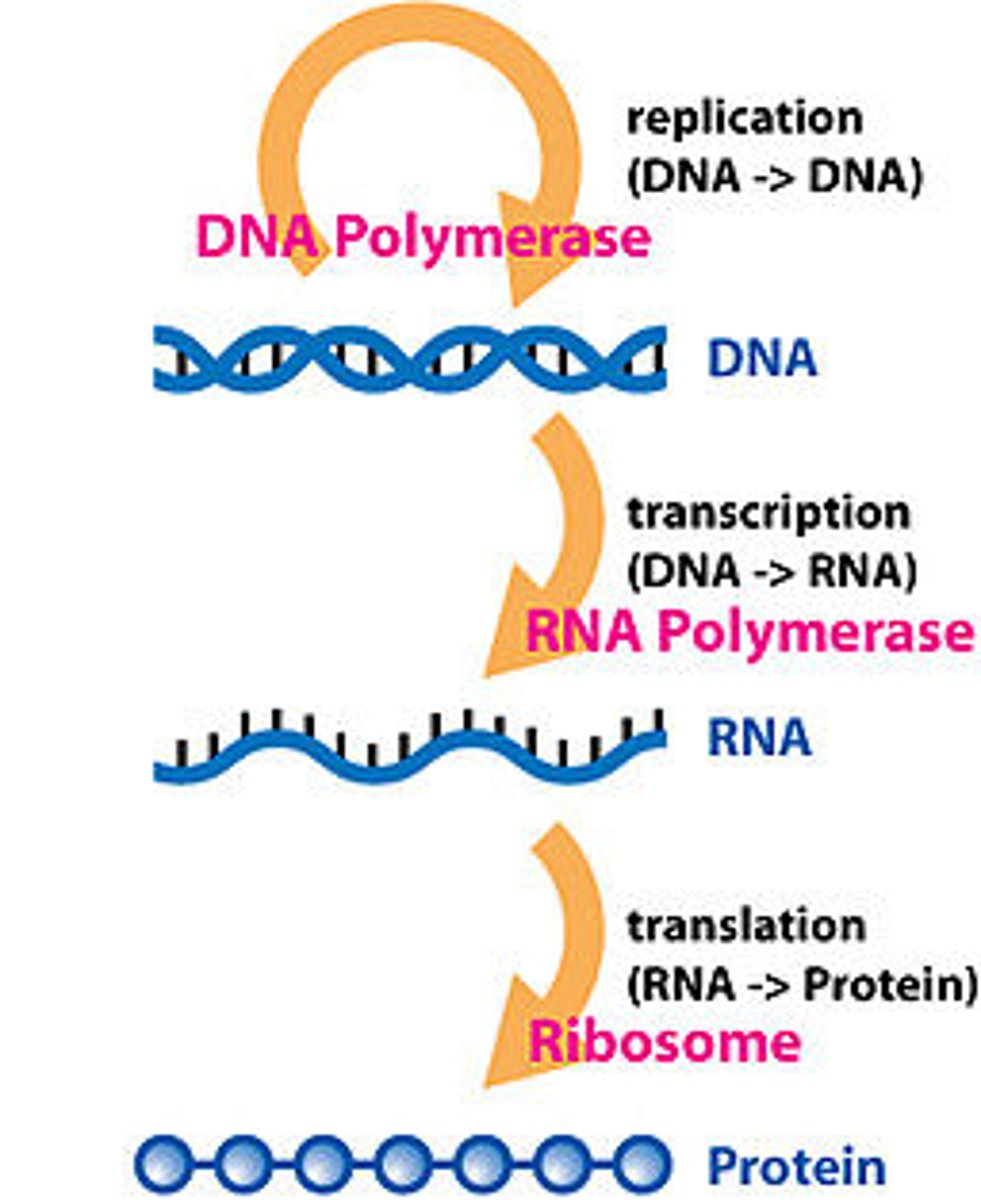
Central dogma
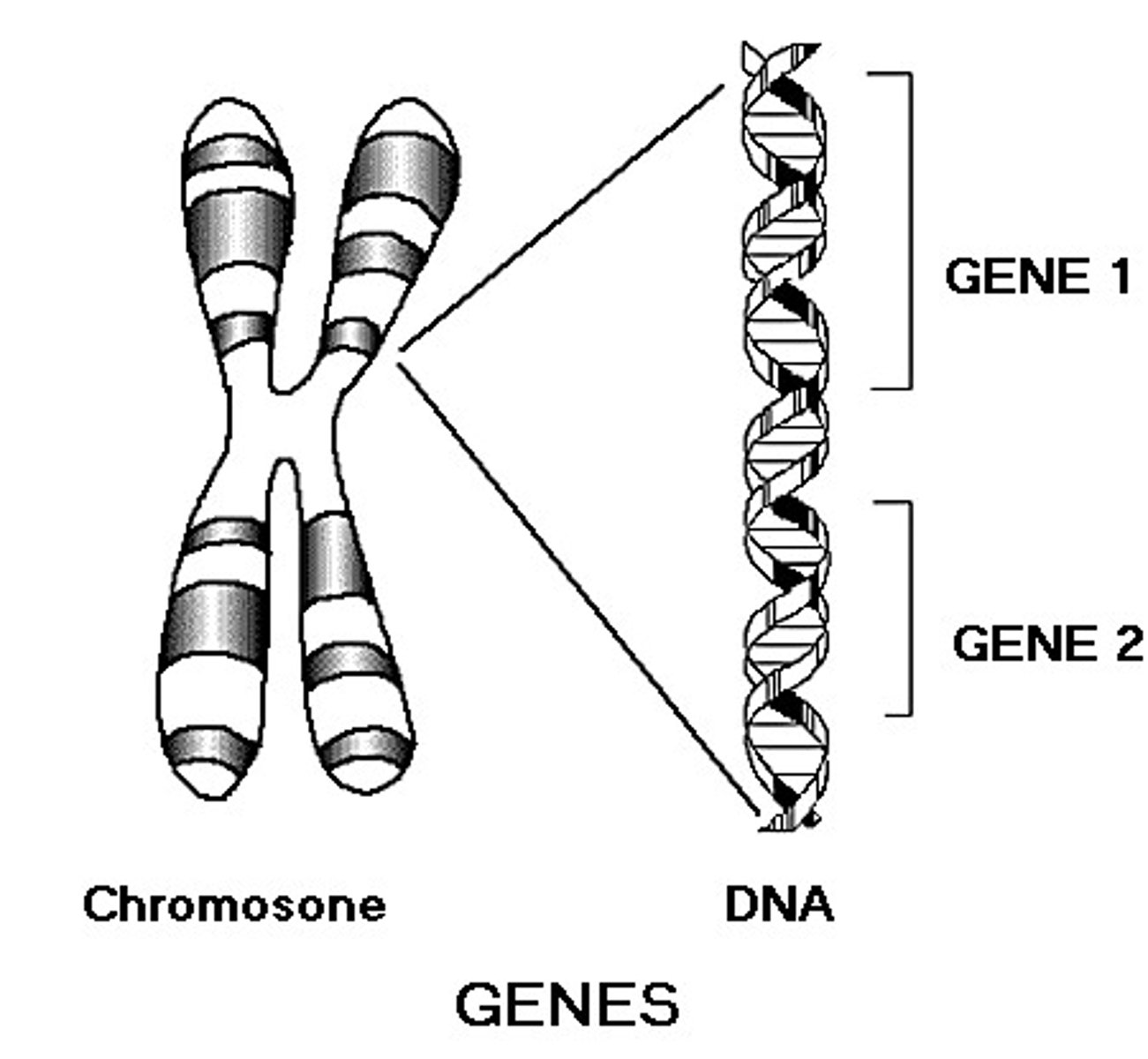
Gene
A unit of DNA that encodes for a specific protein or RNA molecular, which can be expressed via transcription and translation
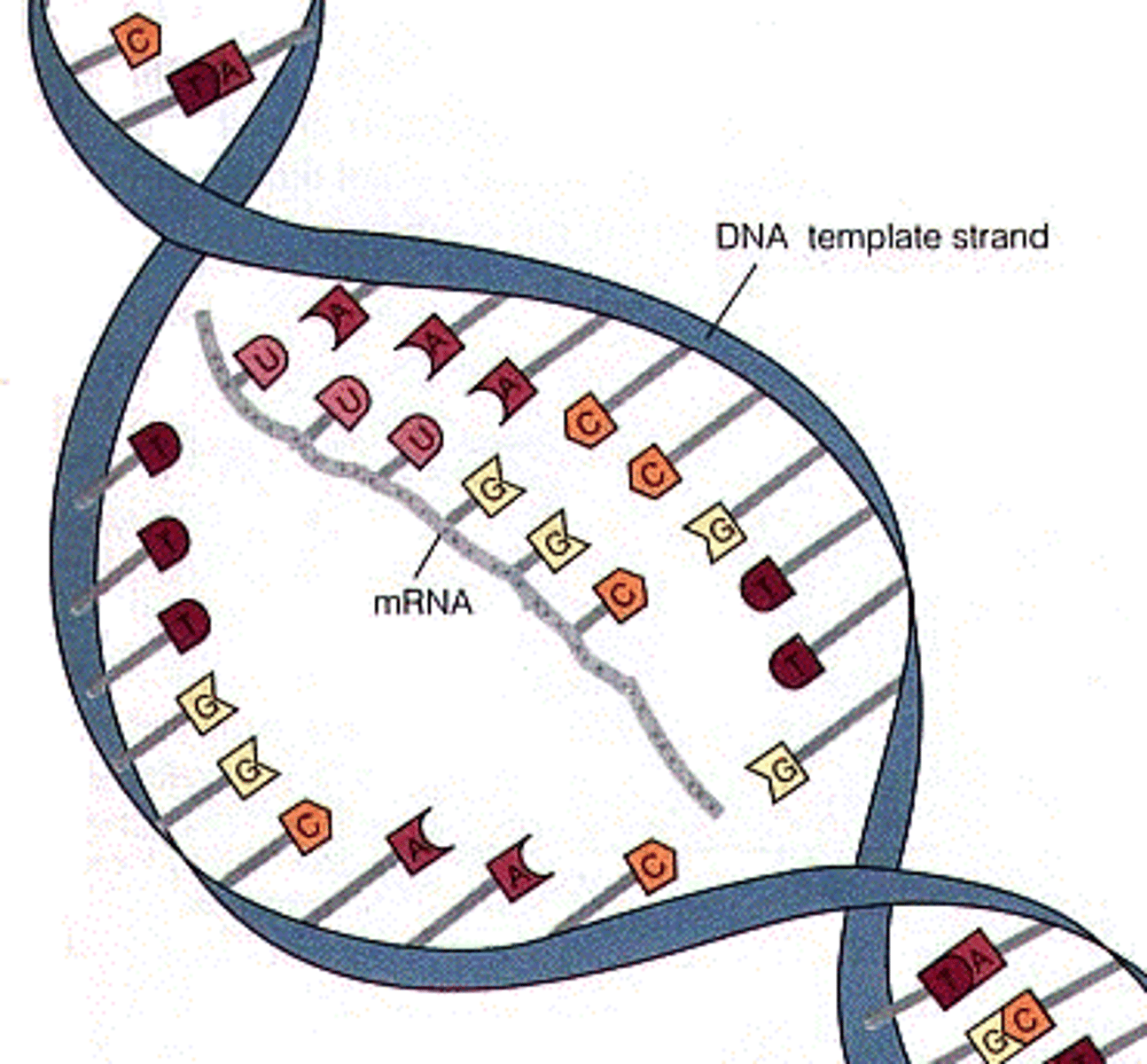
Messenger RNA (mRNA)
Carries the message from DNA in the nucleus via transcription of the gene; it travels into the cytoplasm to be translated.
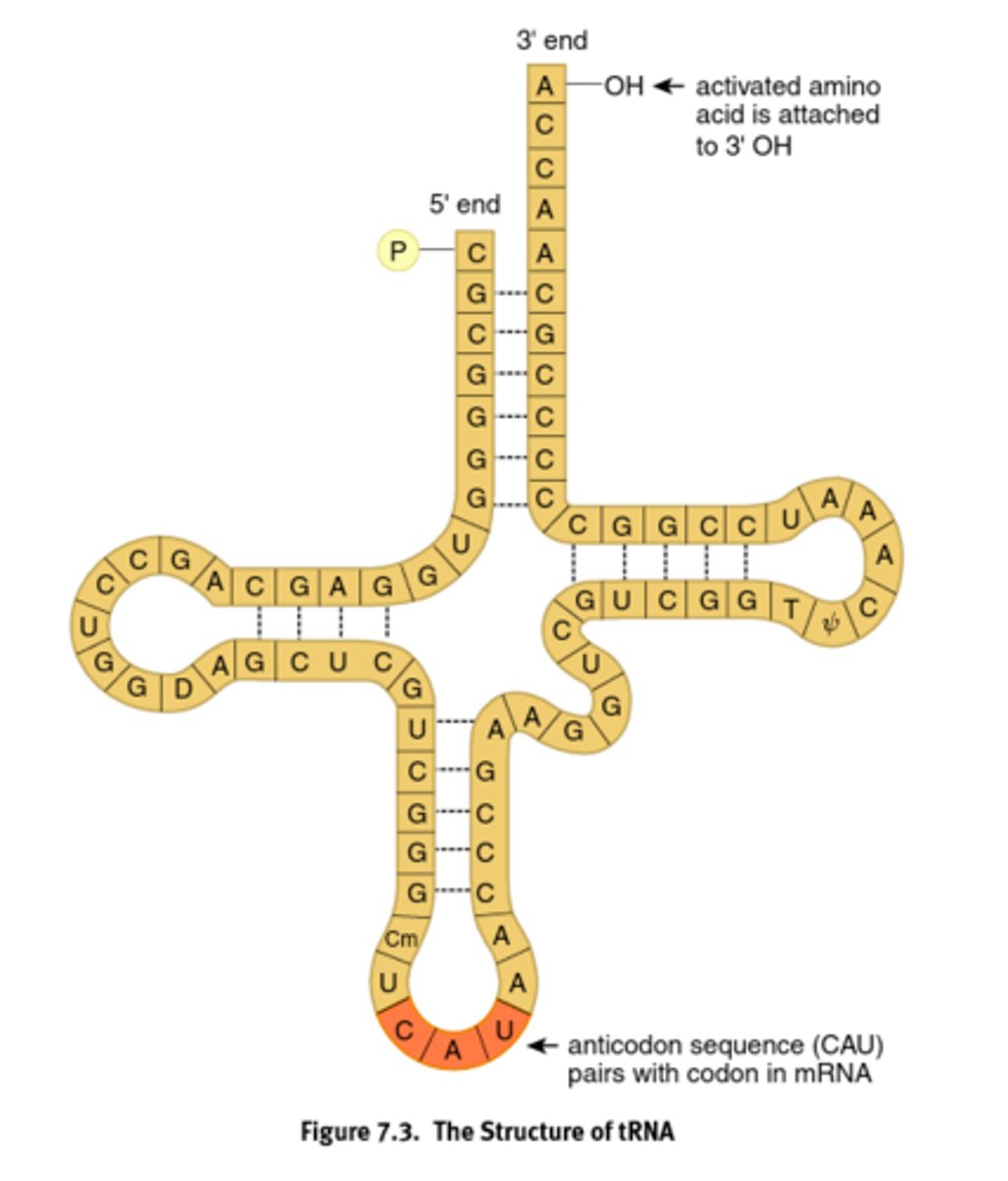
Transfer RNA (rRNA)
Bring the amino acids and recognizes the codon on the mRNA using its anticodon.
When bound to AA, it is charged, and is a high energy bond
Ribosomal RNA (rRNA)
Synthesized in the nucleolus and functions as an integral part of the ribosomal machinery used during protein assembly in the cytoplasm.
Makes up the ribosome and its enzymatically active.
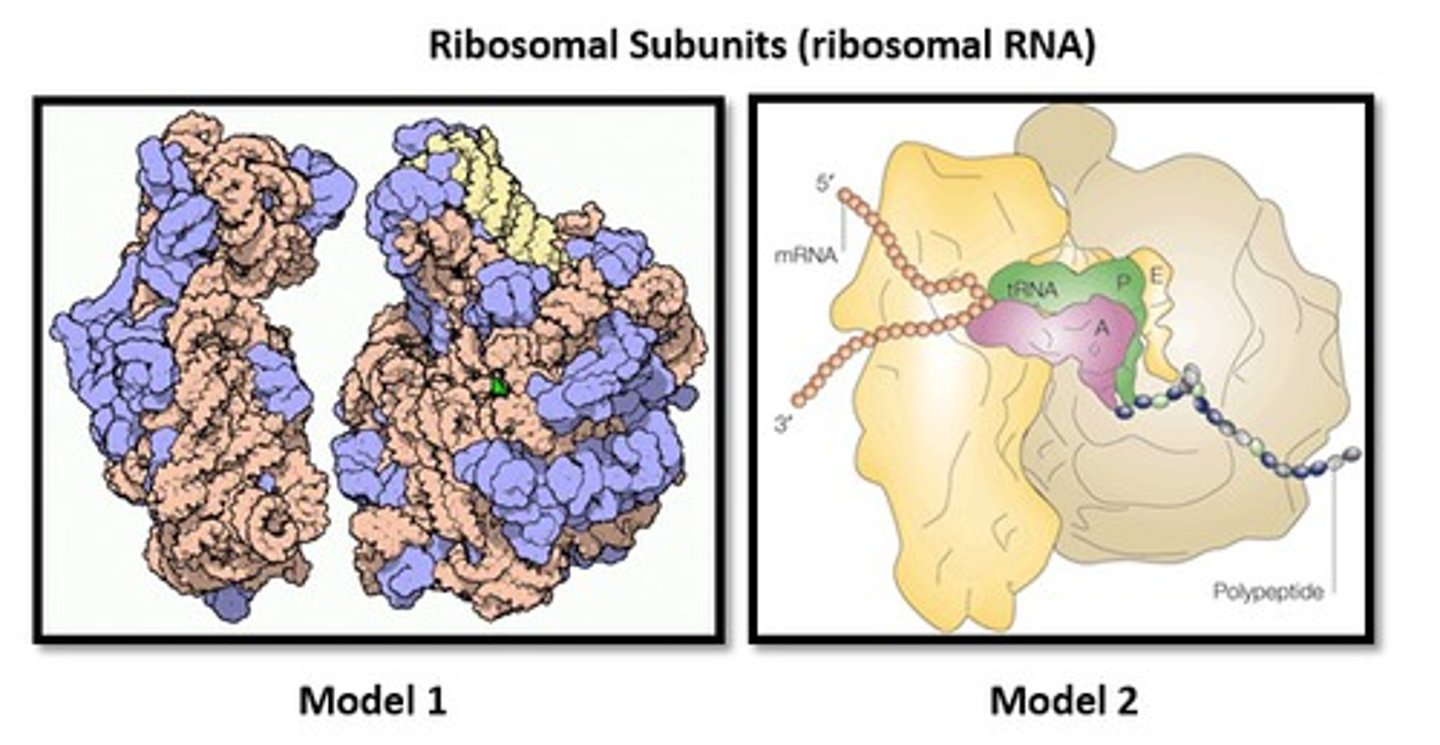
ribozyme
a type of RNA that can act as an enzyme
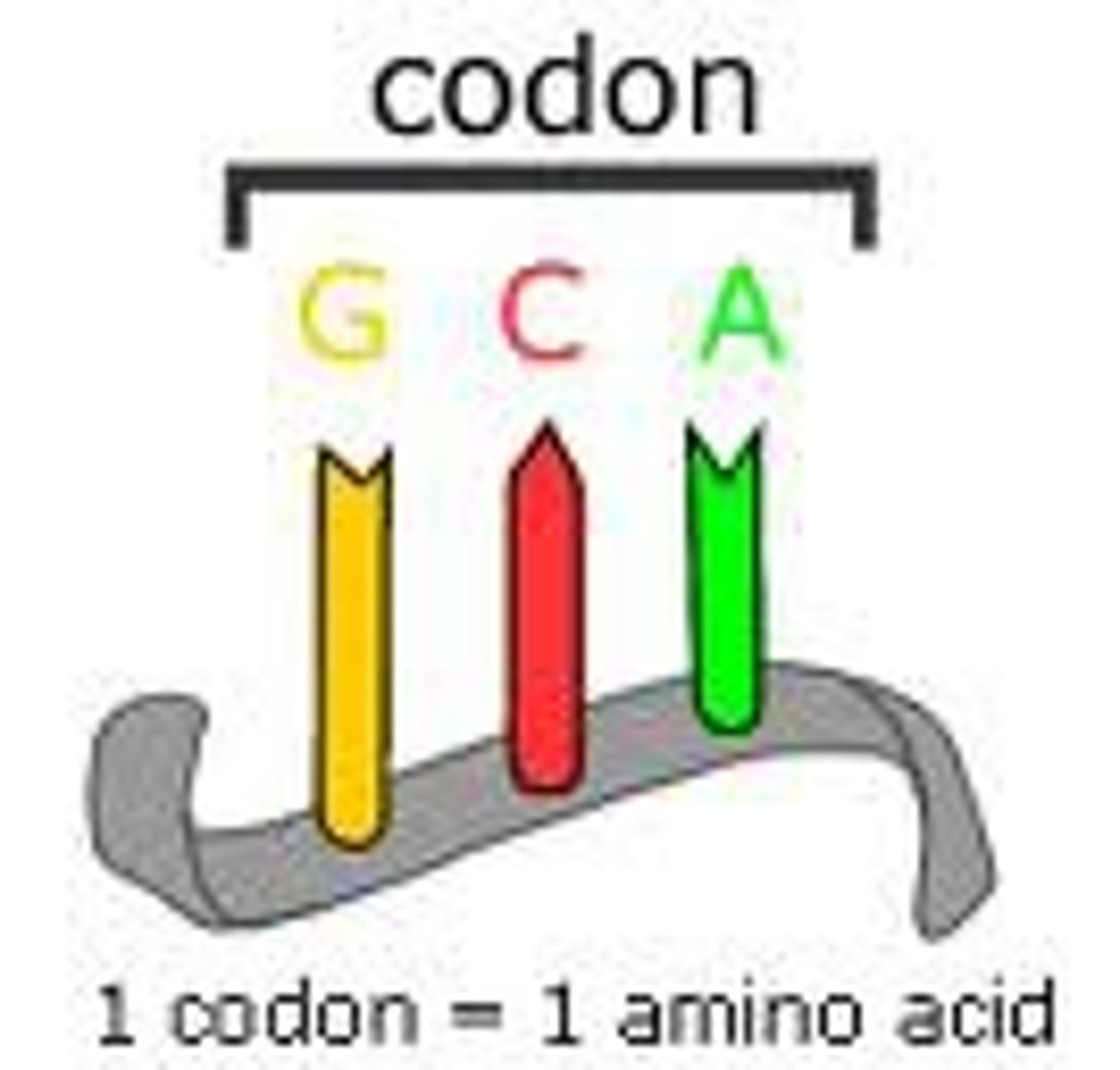
Codon
A three-nucleotide sequence of DNA or mRNA that specifies a particular amino acid or termination signal; the basic unit of the genetic code.
Start codon
AUG
think starts school in AUGest
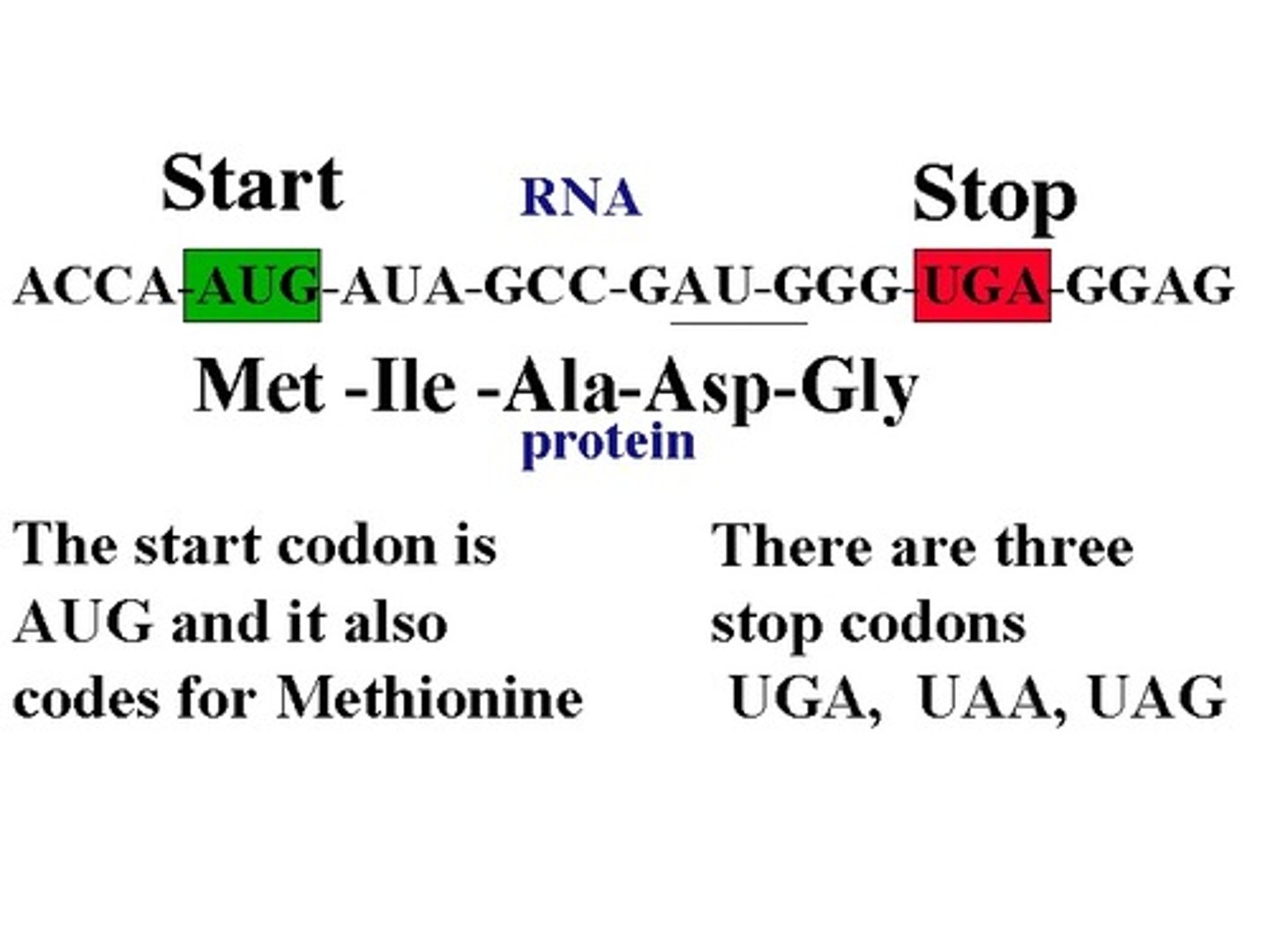
Stop codons
1) UGA - U Go Away
2) UAG - U Are Gone
3) UAA - U Are Annoying

anticodon
group of three bases on a tRNA molecule that are complementary to an mRNA codon
Point mutations
Specific mutations one piece of a codon:
1) Silent
2) Nonsense
3) Missense
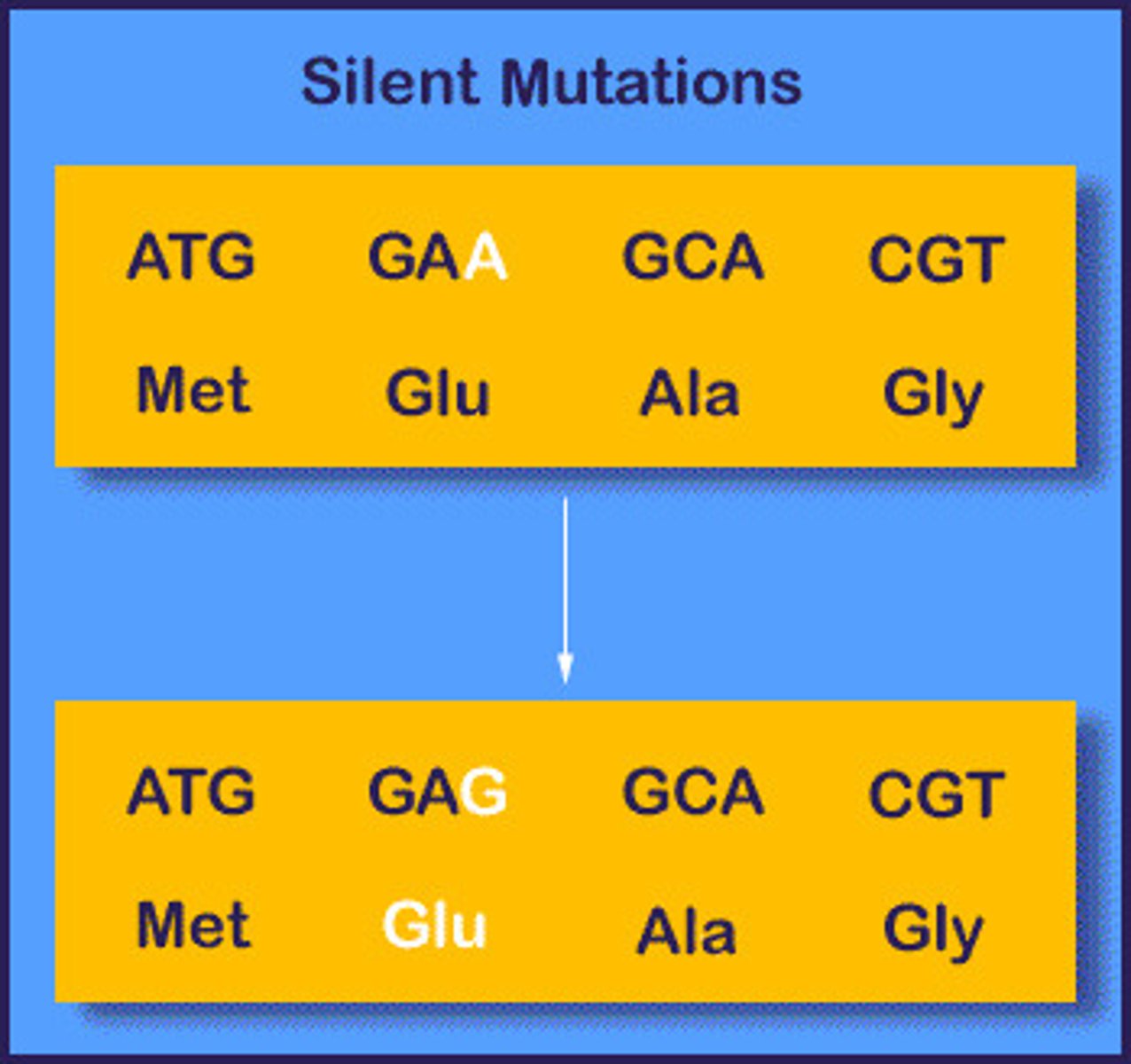
Silent mutations
Causes no effect in the protein sequence due to redundancy in the wobble position (third base in codon)
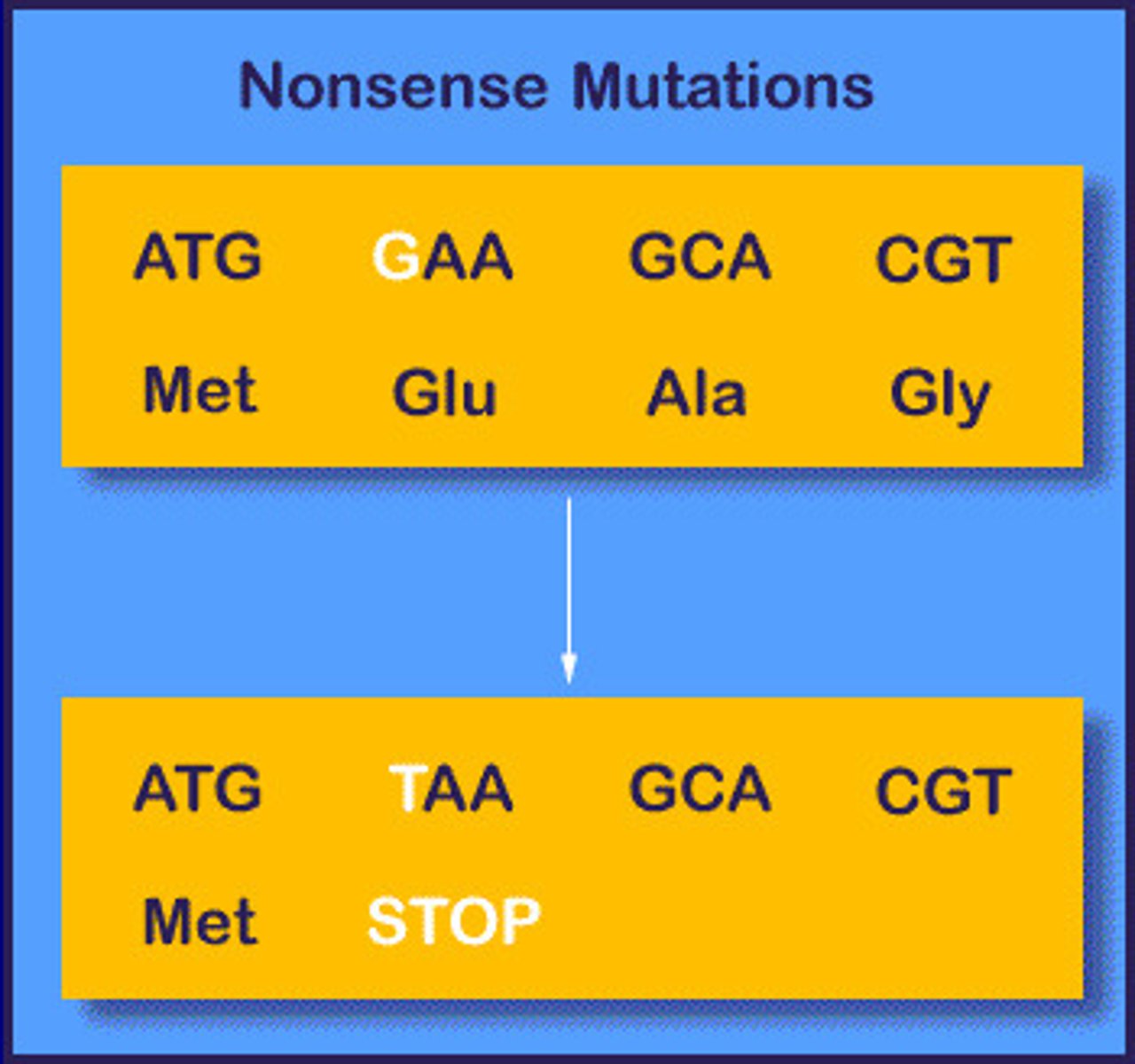
Nonsense (truncation)
Mutation that produces a premature stop codon.
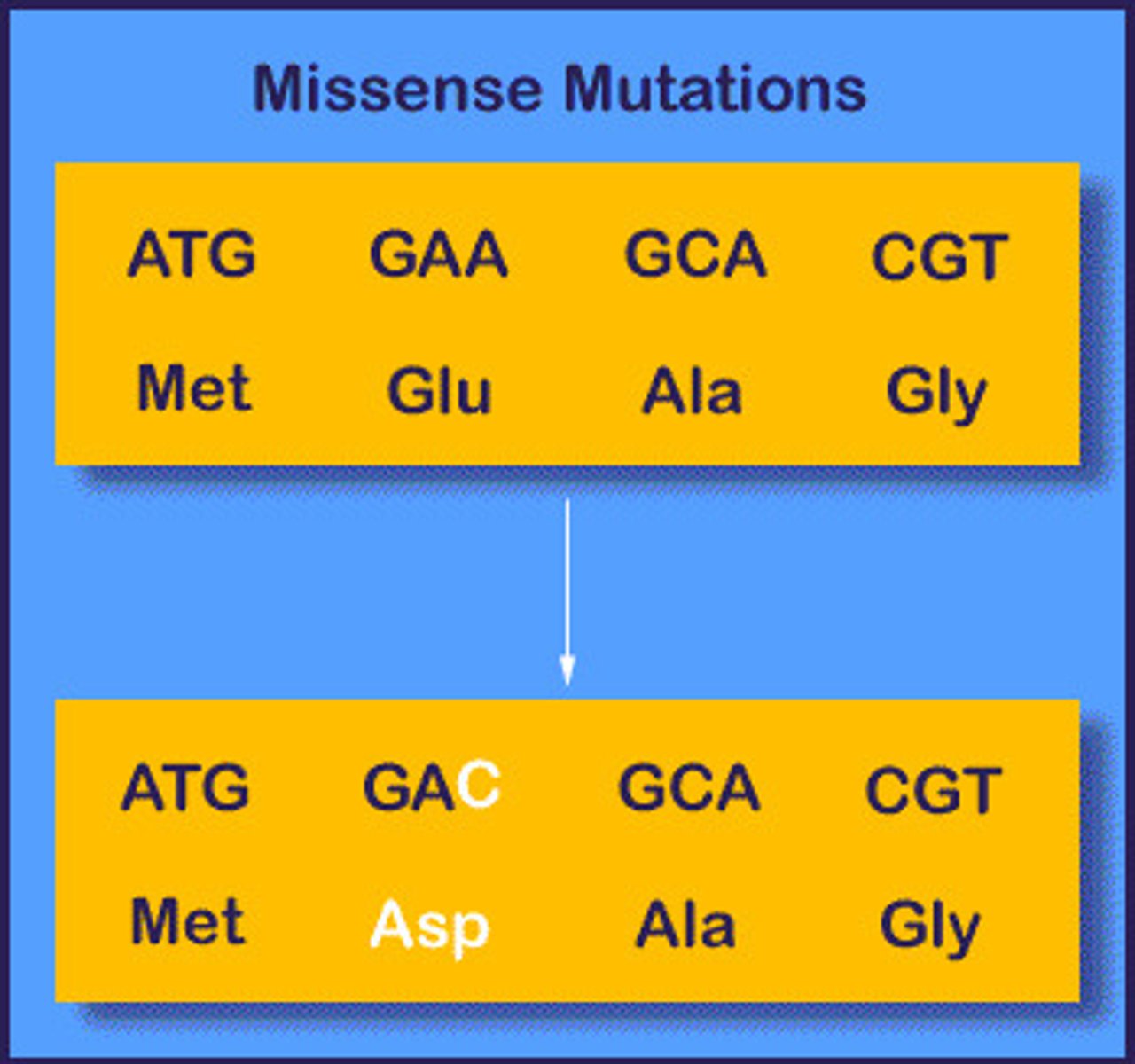
Missense mutation
Mutation that produces a codon that codes for a different amino acid.
Frameshift mutation
Mutation that results from nucleotide addition or deletion, and change the reading frame of subsequent codons.
RNA vs DNA
RNA has:
1) A ribose sugar instead of deoxyribose
2) Substitution of uracil for thymine
3) Single-stranded instead of double-stranded
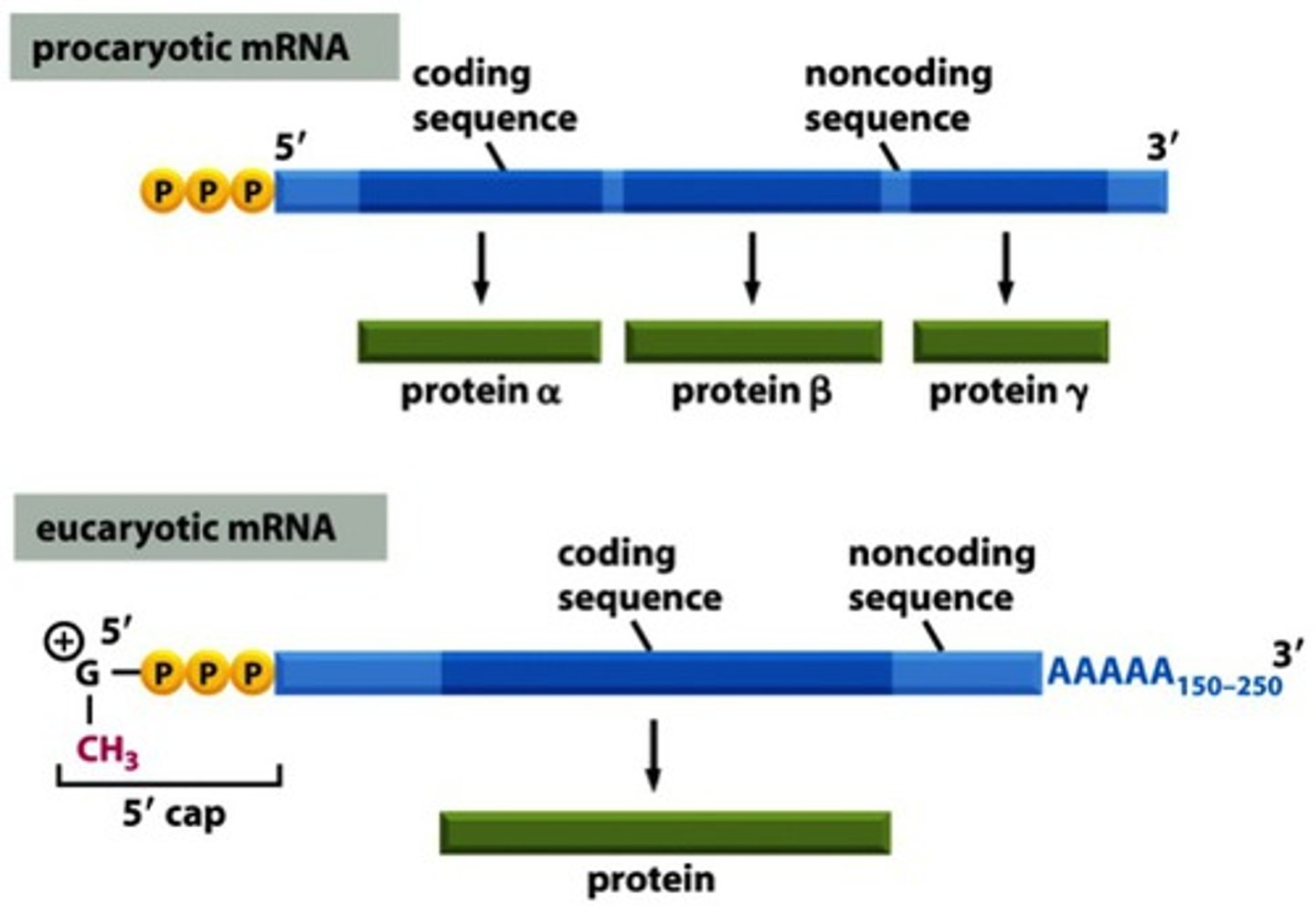
Monocistronic
The coding pattern of eukaryotes in which one mRNA molecule codes for only one protein.
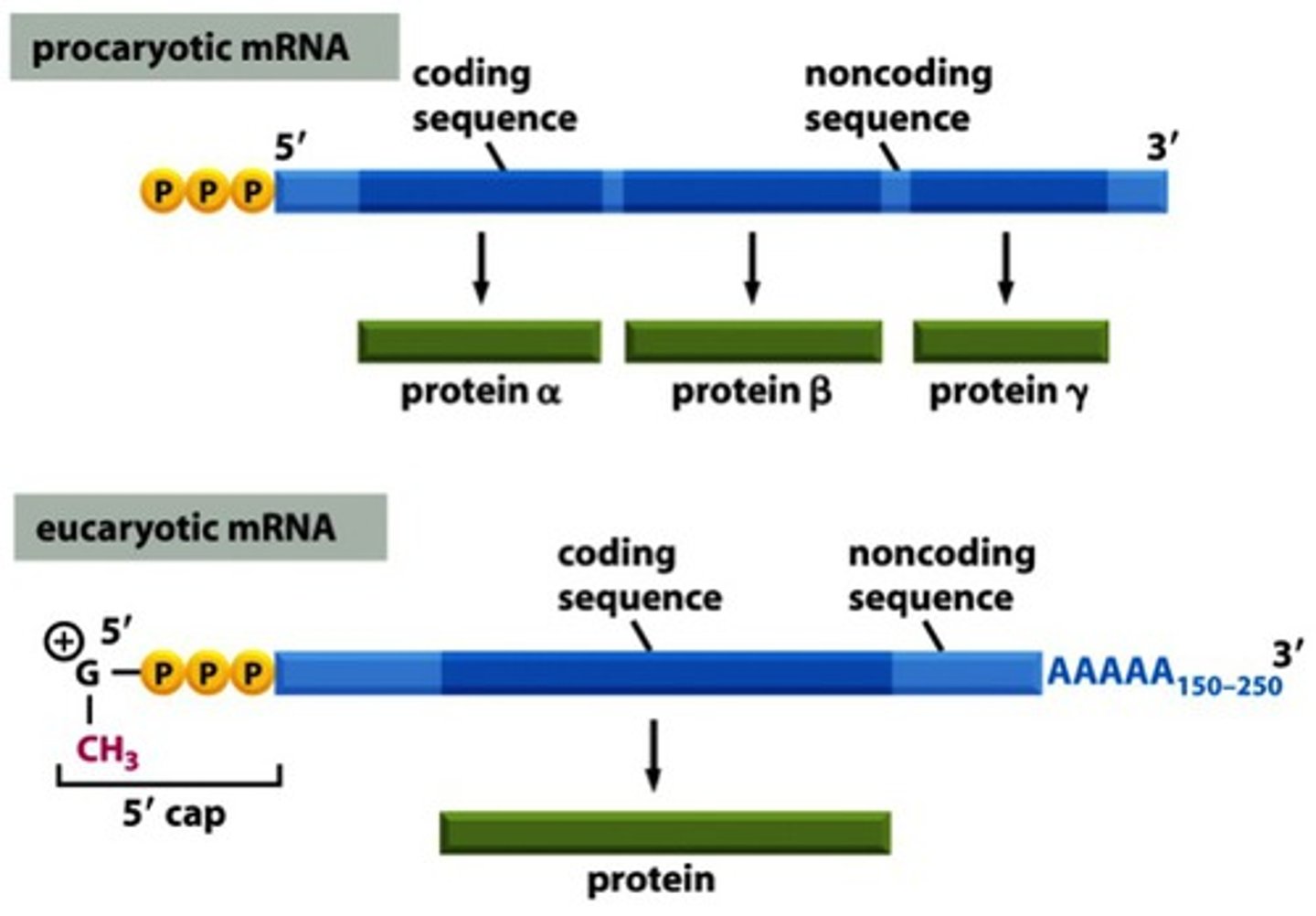
Polycistronic
The coding pattern of prokaryotes, in which one mRNA may code for multiple proteins.
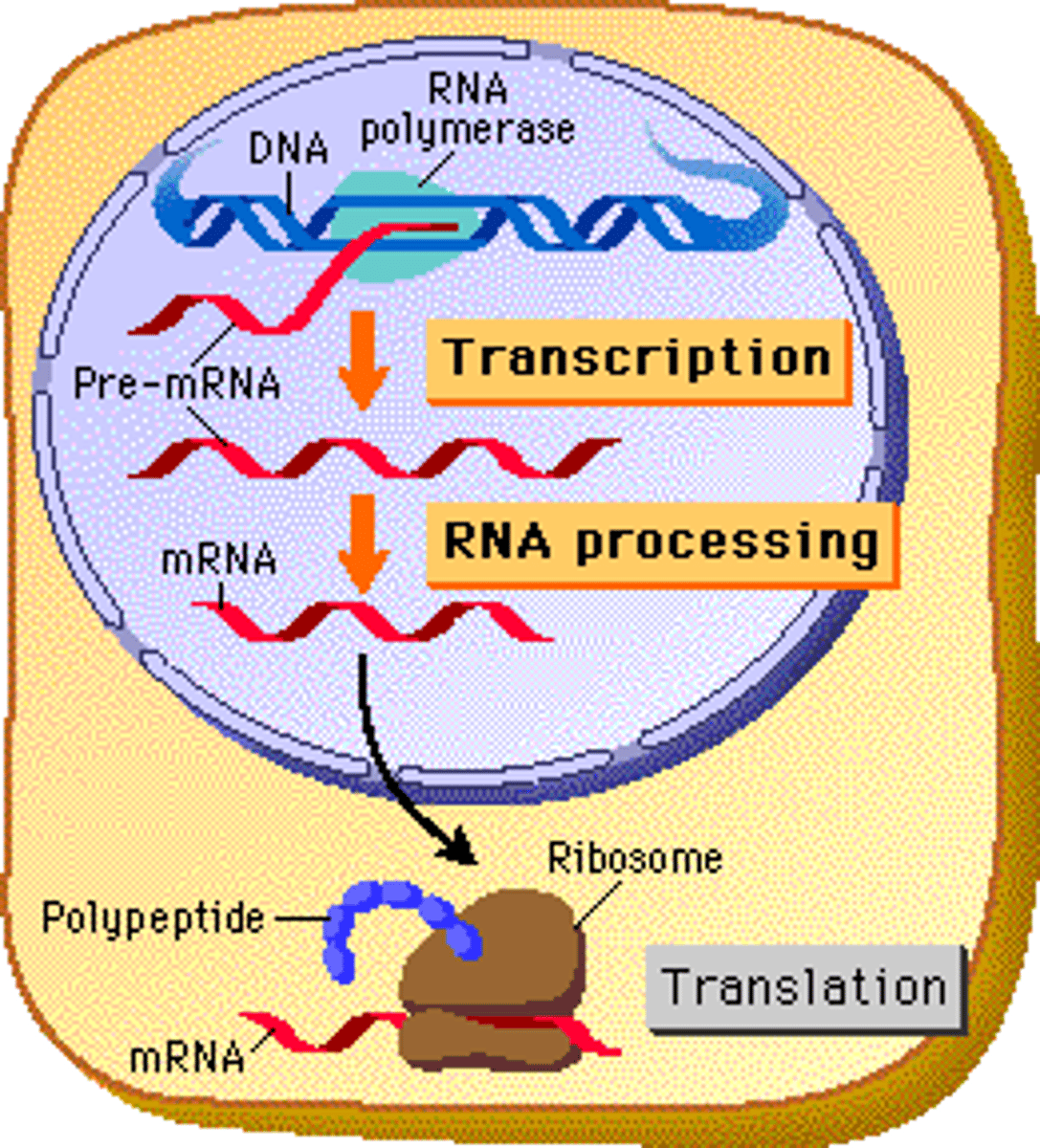
Transcription
The creation of mRNA from a DNA template.
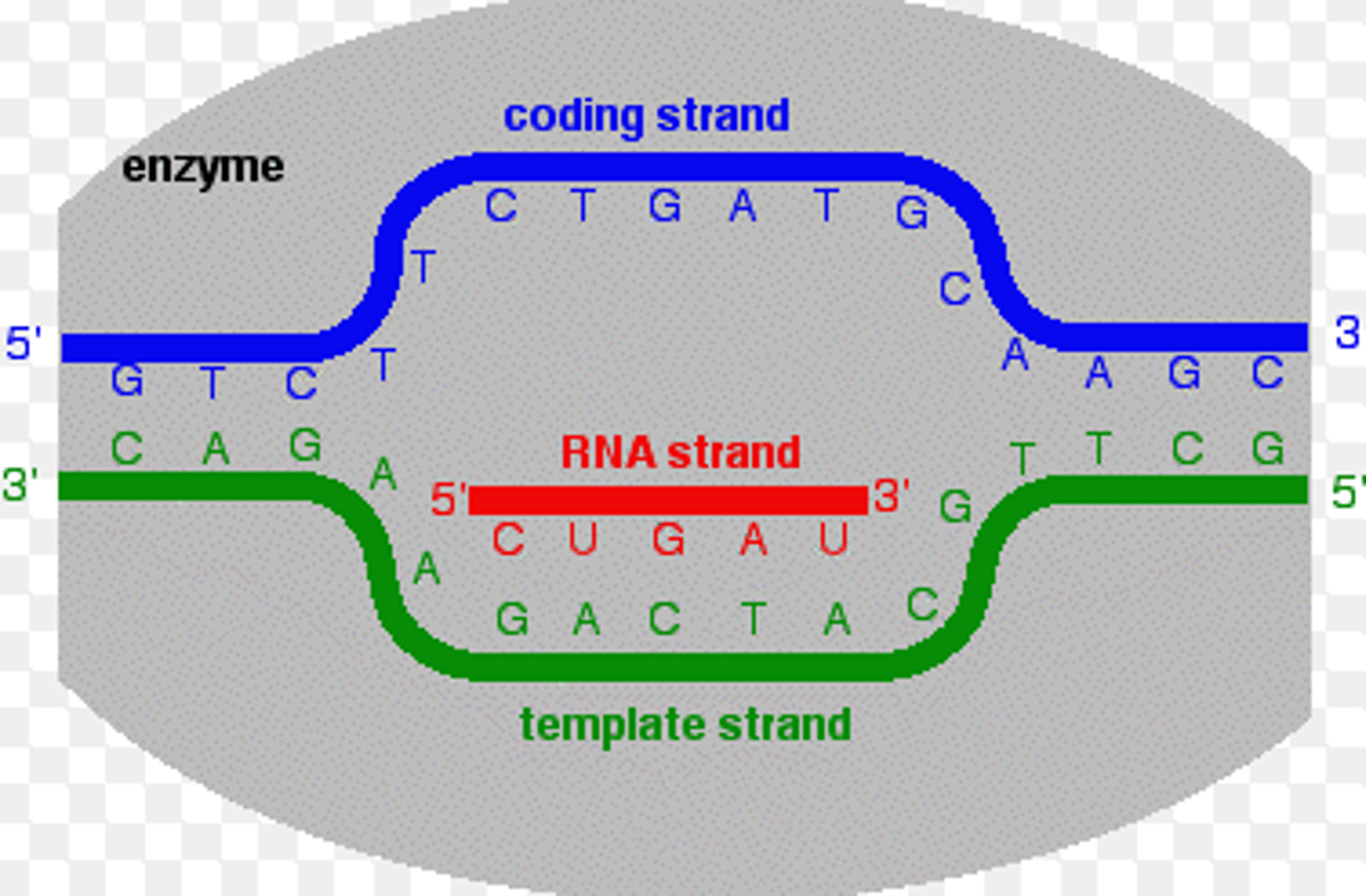
DNA strands to be transcribed
1) Coding strand
2) Template strand
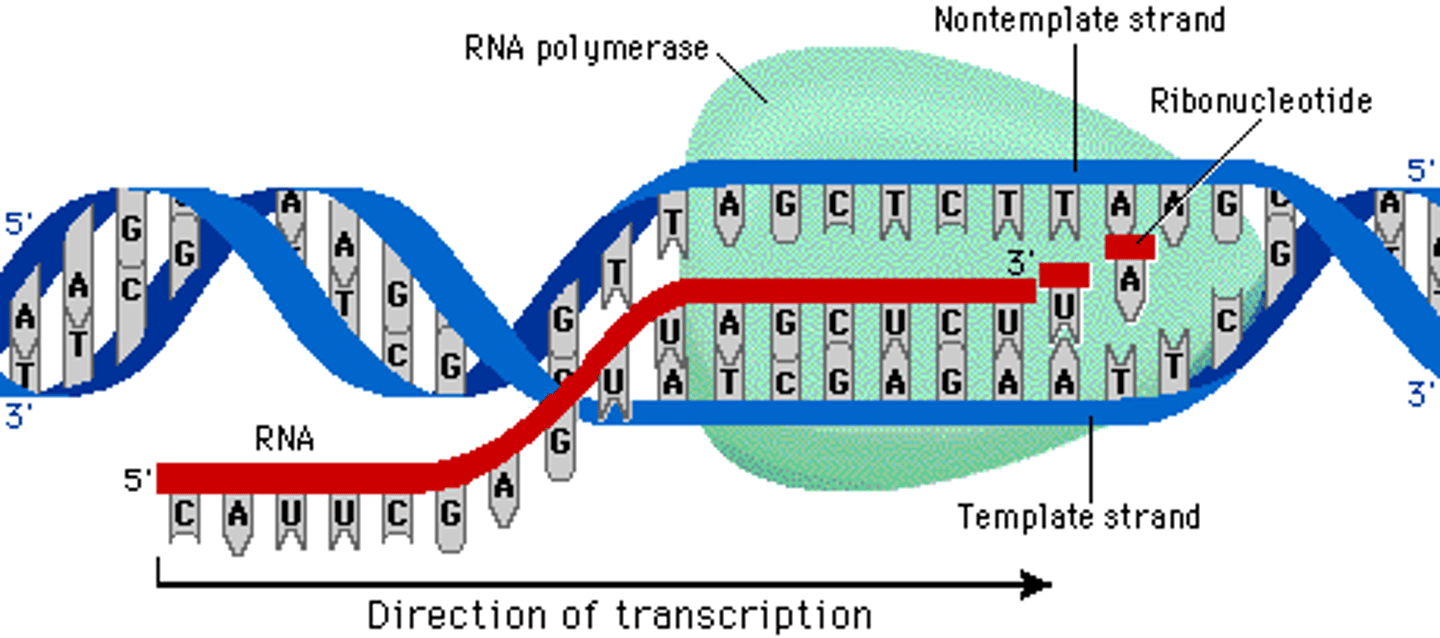
RNA polymerase
Enzyme that links together the growing chain of RNA nucleotides during transcription using a DNA strand as a template
1) Reads 3' to 5' but adds 5' to 3'
2) Does no proof reading
3) Continues until it reaches a termination signal.
hnRNA
Heterogeneous RNA.
RNA that has been freshly transcribed and has not underwent any post transcriptional modifications.
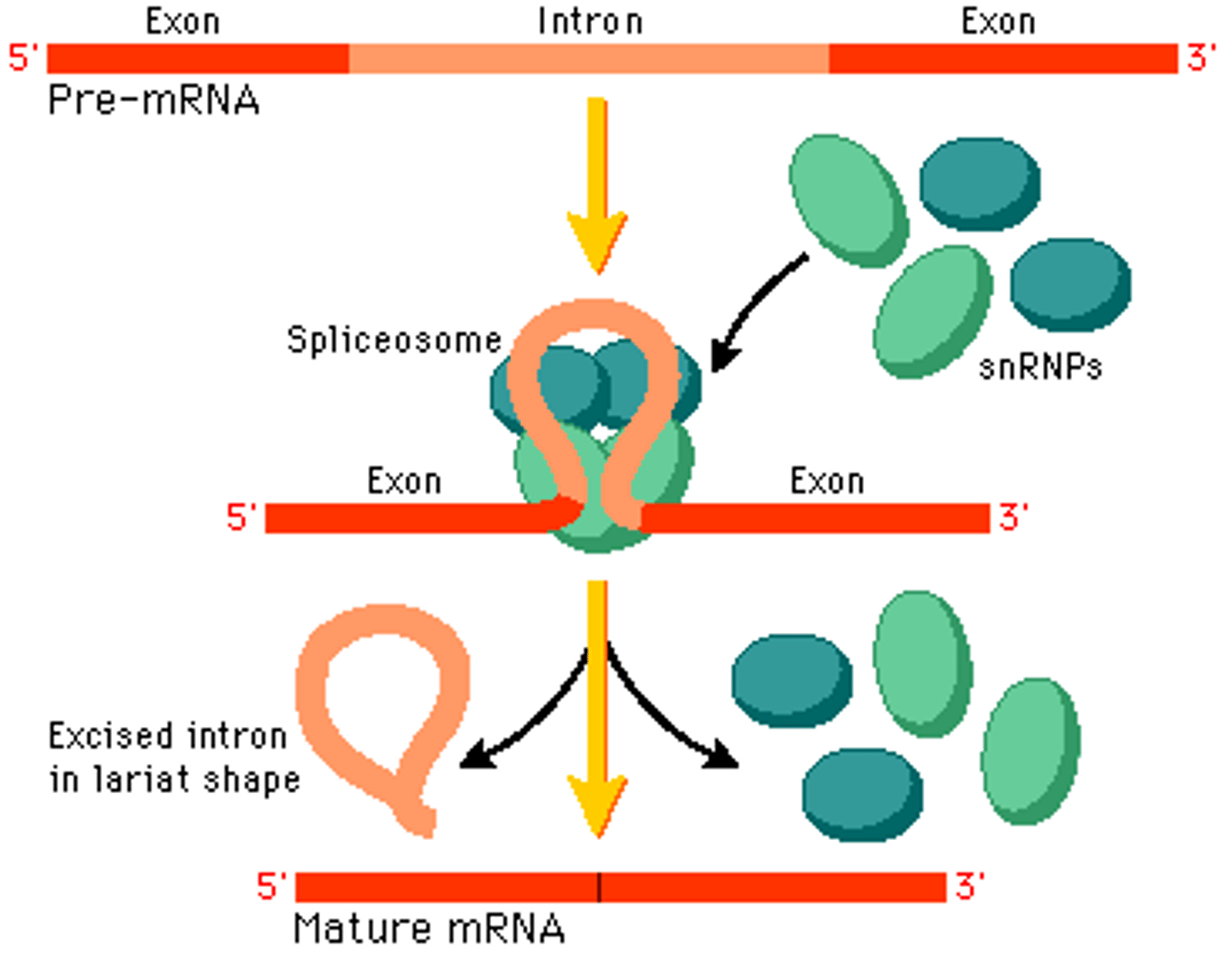
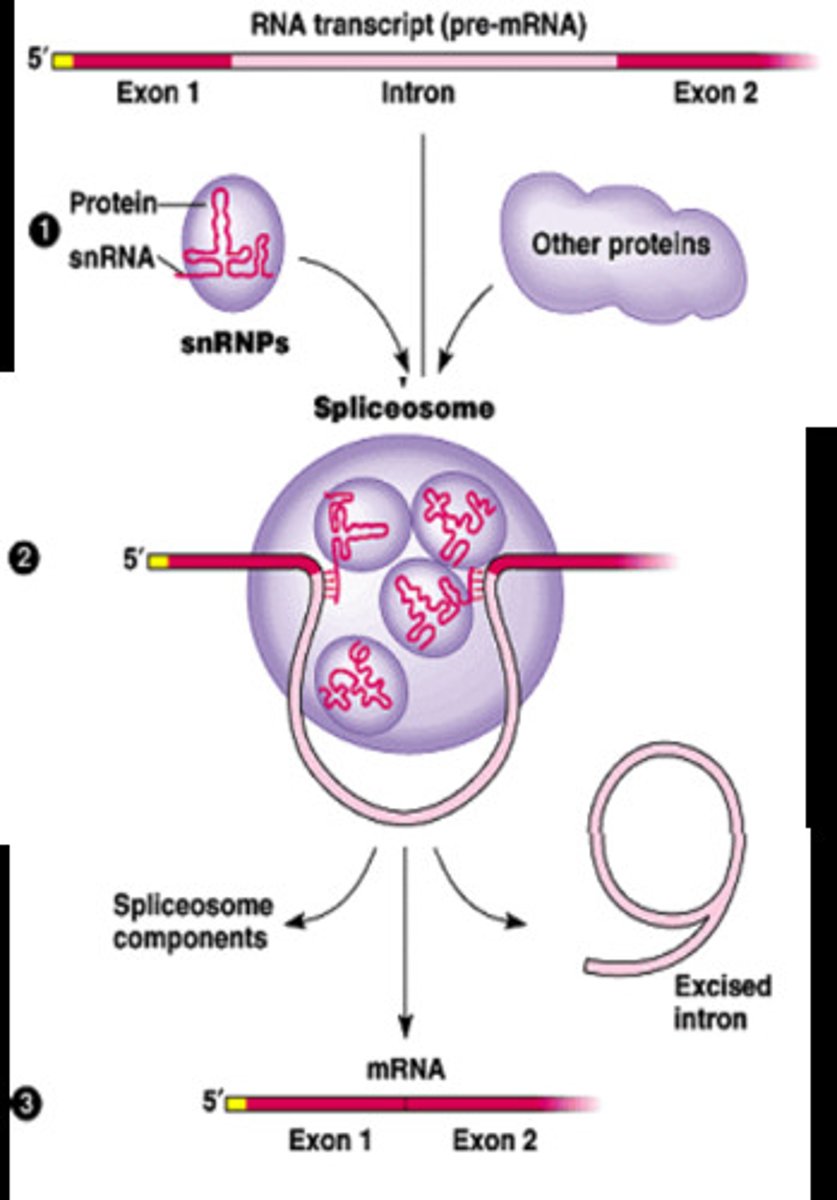
Splicesome
a complex of specialized RNA and protein subunits that removes introns from a transcribed pre-mRNA segment.
Contain:
1) snRNA
2) snRNPs or "snurps"
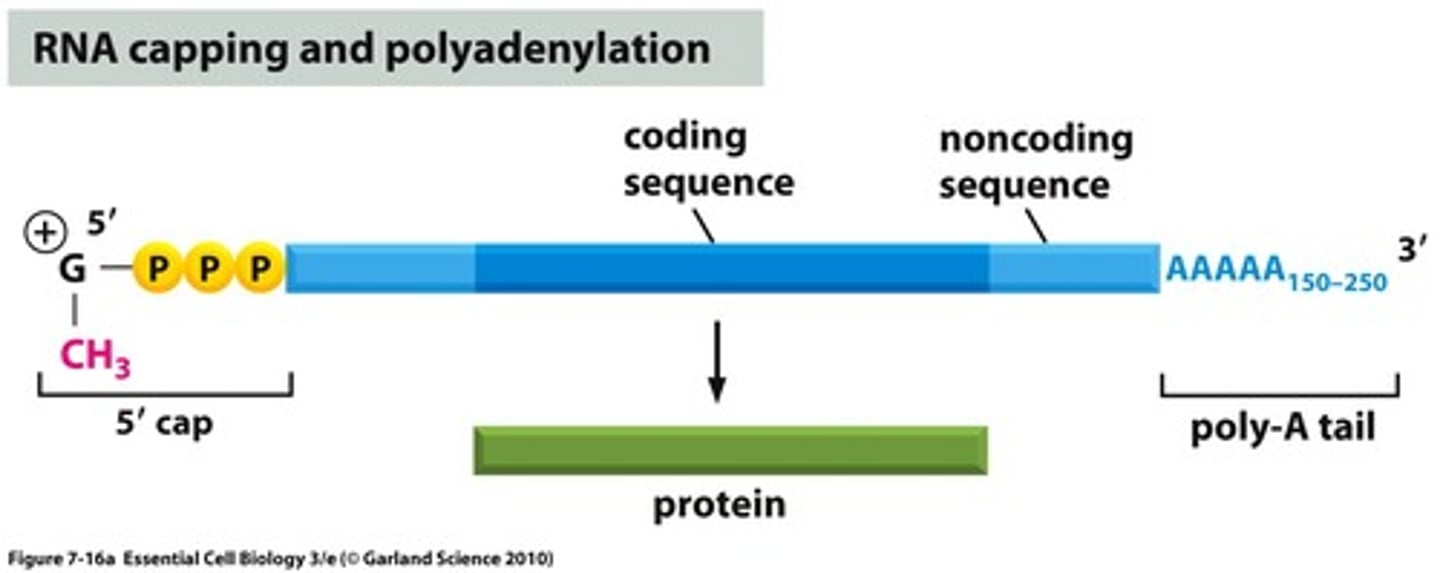
5' Cap
A 7-methylguanylate triphosphate cap that protects mRNA from degradation in cytoplasm and is a recognized binding site for ribosomes.
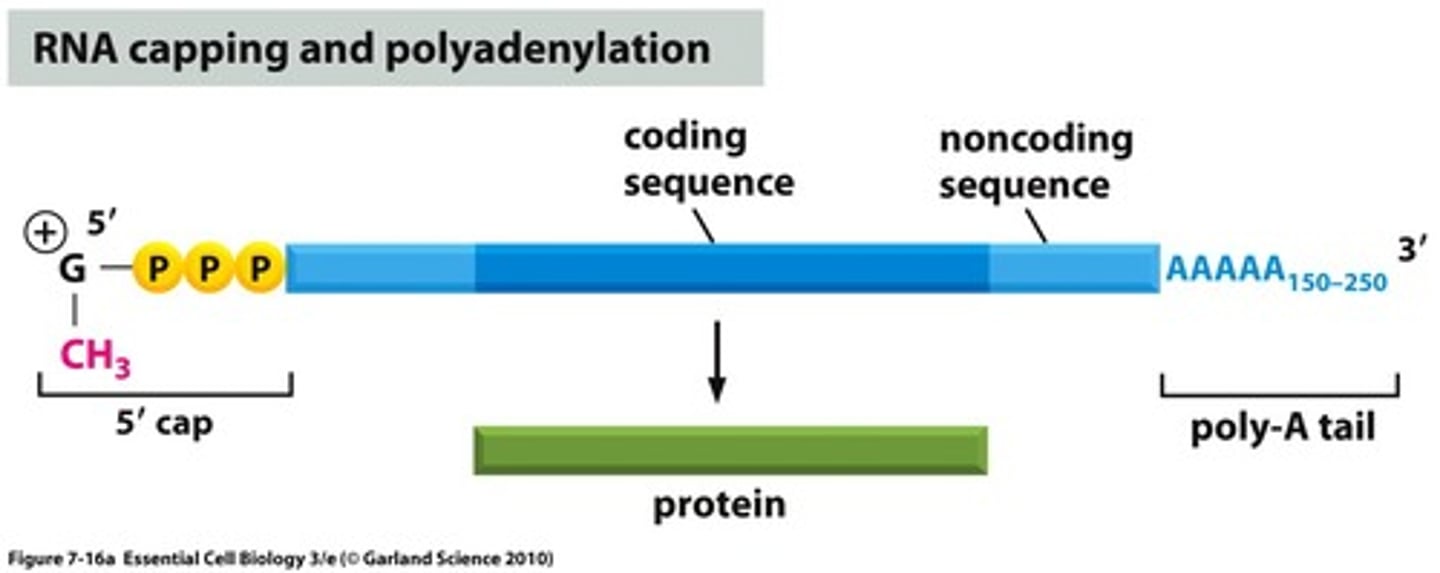
3' Poly-A tail
A poly-A tail is composed of adenine and is added to the 3' end of the mRNA transcript and protects the message against rapid degradation.
Longer the poly-A tail = longer time before degradation.
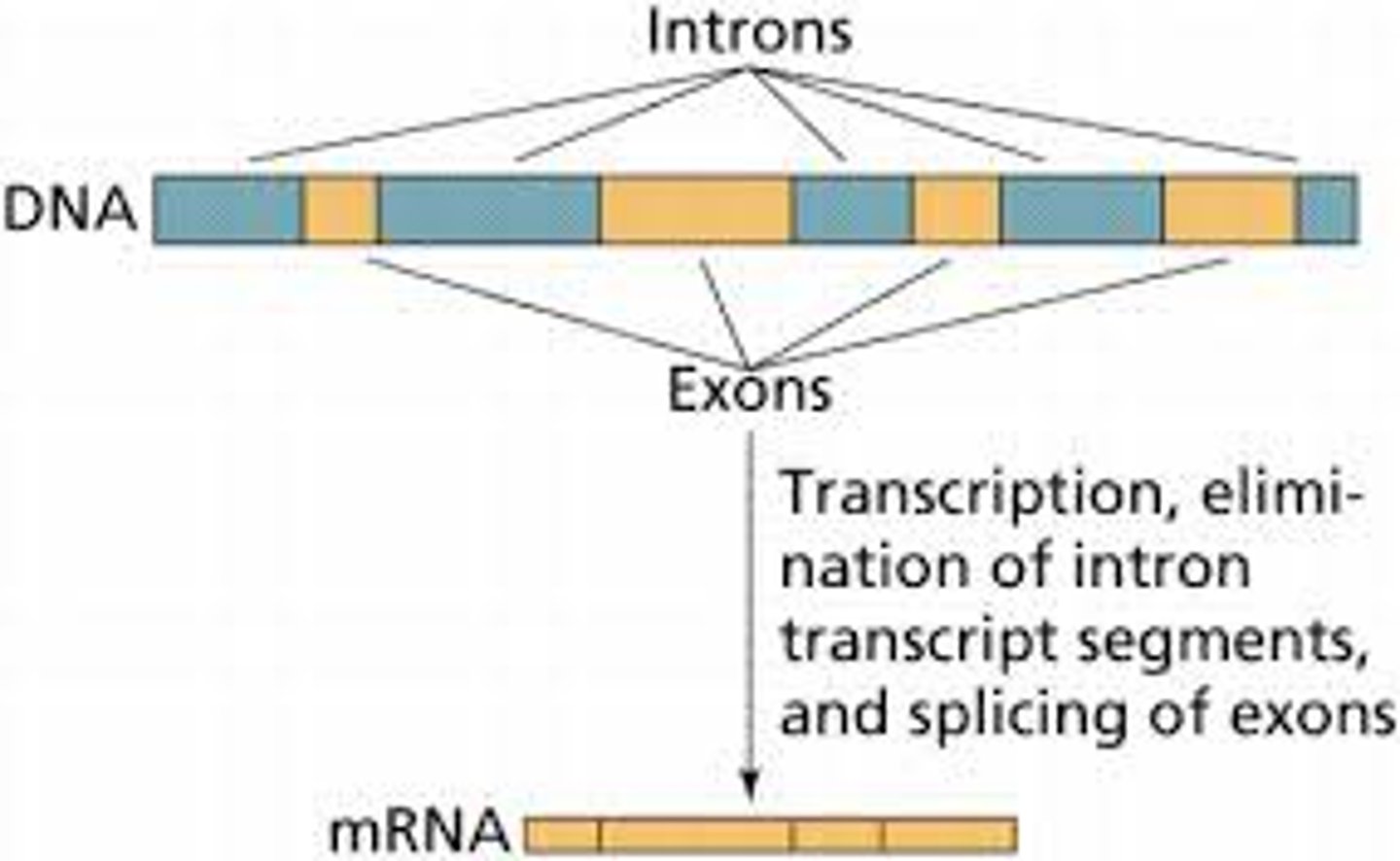
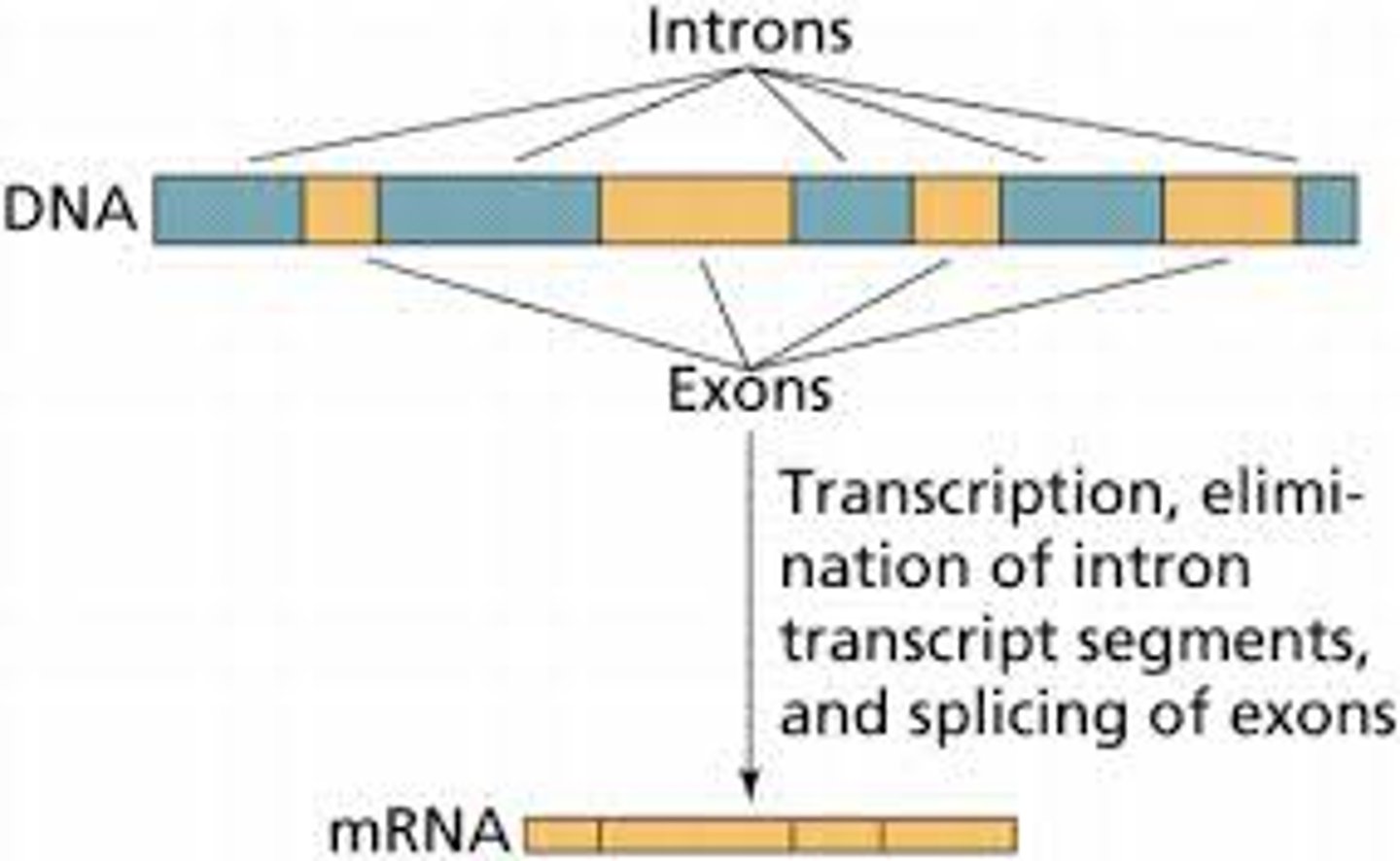
Introns
Noncoding segments of nucleic acid that lie between coding sequences.
Mechanism of transcription
1) Helicase and topoisomerase unwind the dsDNA.
2) RNA polymerase II binds to TATA box within the promoter region of the gene (25 base pairs upstream)
3) hnRNA is synthesized from the DNA template (antisense) strand and stopped once termination signal.
Post-transcriptional modifications
4) A 7-methylguanylate triphosphate cap added to 5' end.
5) Poly-A tail added to 3' end
6) Splicing done by snRNA and snRNPs in the spliceosome. Process removes introns.
7) Alternative splicing (combining of exons) allows more variability of gene products
8) Leave nucleus and into cytosol.
Alternative splicing
Splicing of introns in a pre-mRNA that occurs in different ways, leading to different mRNAs that code for different proteins or protein isoforms. Increases the diversity of proteins.
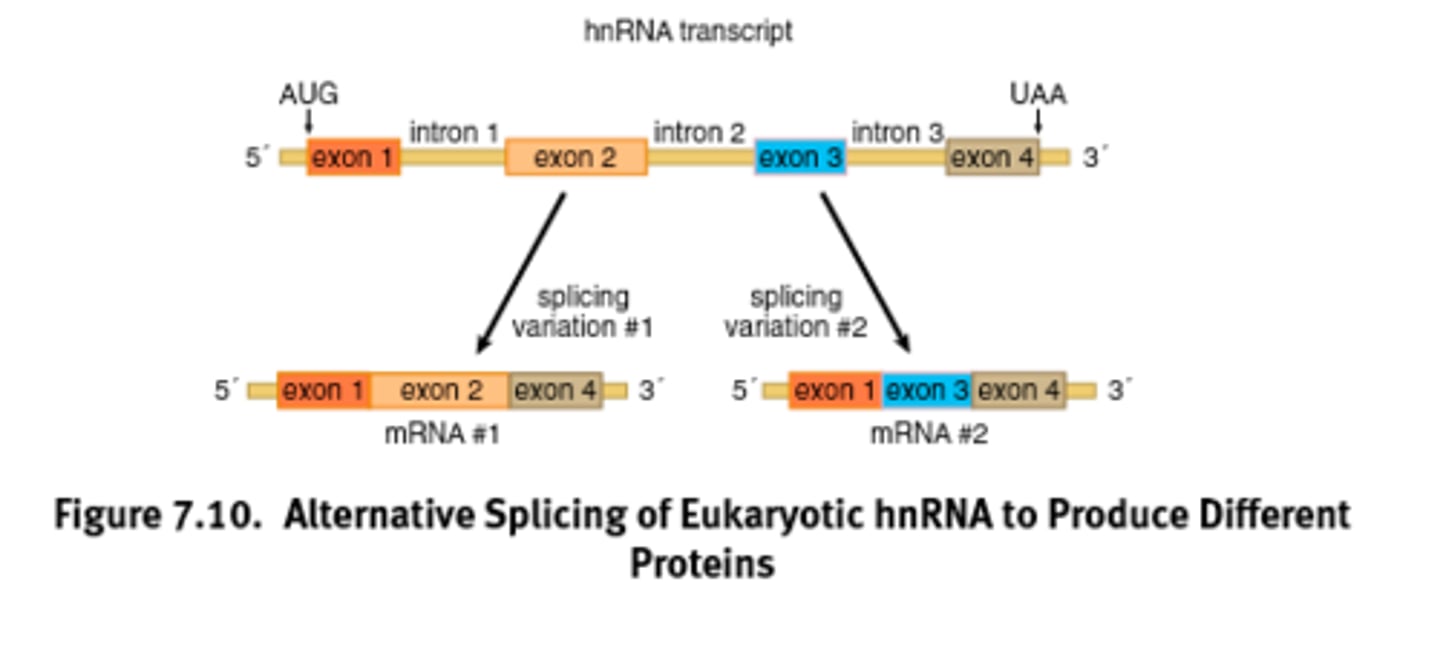
Exons
A coding region of a eukaryotic gene. Exons, which are expressed, are separated from each other by introns.
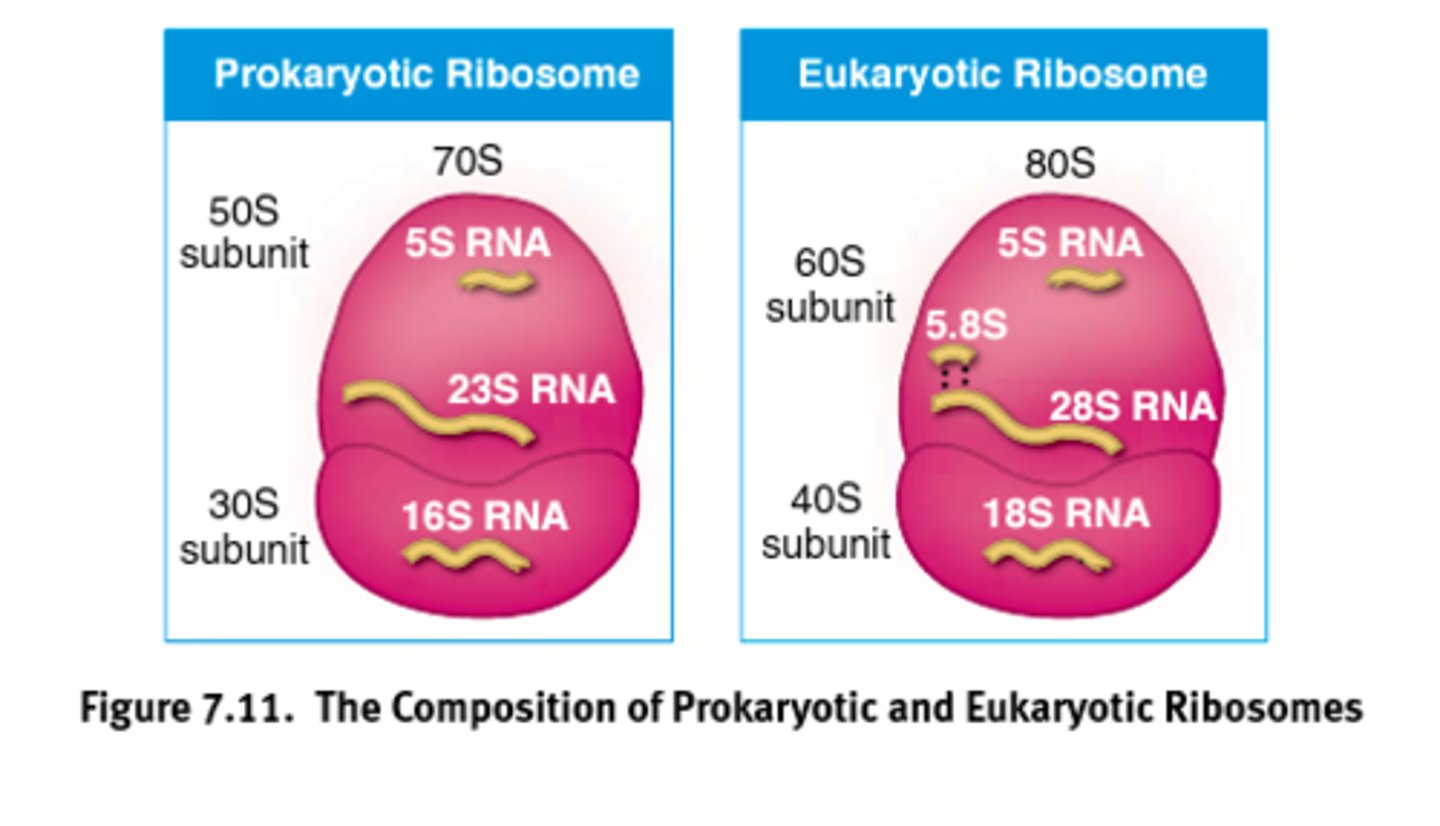
Ribosome
Factories for the translation of mRNA into proteins.
Composed of proteins and rRNA with variability among prokaryotes and eukaryotes.
Ribosomal sub-weight
(Two domains)
Eukaryotic Ribosome = 80 S (60+40)
Prokaryotic Ribosome = 70 S (50+30)
Where S value is determined experimentally by studying the behavior of particles in a centrifuge.
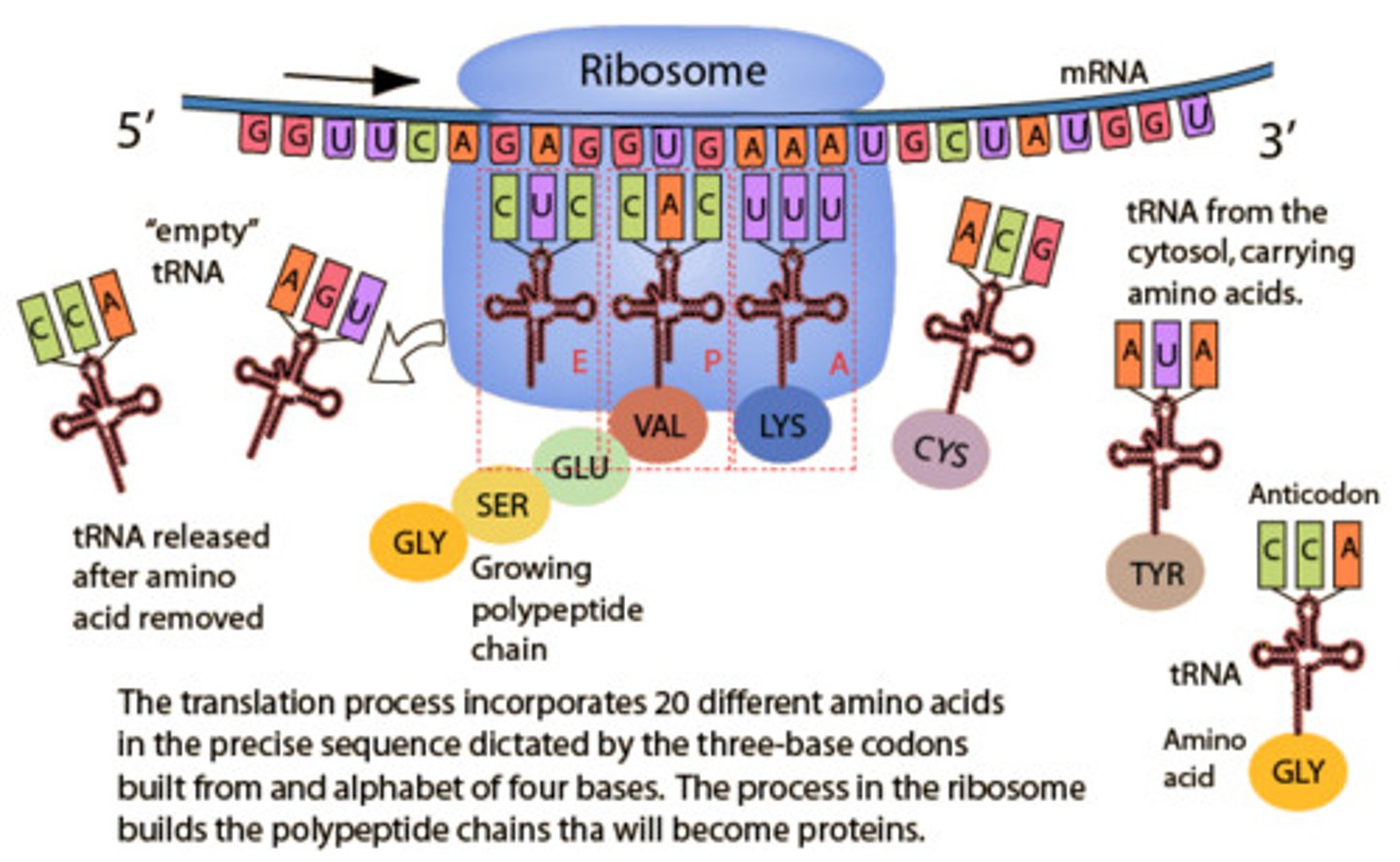
Mechanisms of Translation
1) Initiation begins for eukaryotes when 40S ribosome attaches to 5' cap and begins scanning for a start codon.
2) Once AUG is found, it lays down a methionine in the P site of the ribosome.
3) Elongation begins and new tRNA come into site A of the ribosome and transfer growing polypeptide chain from tRNA in the P site to tRNA in the A site.
4) The now uncharged tRNA exits the ribosome via the E site.
5) Process continues until stop codon is reached and a release factor places a water molecule on the polypeptide chain causing release of protein.
Post-translational modifications
6) Folding by chaperones, formation of quaternary structure, cleavage of protein or signal sequences, or the covalent addition of other biomolecules.
Phosphorylation
The addition of phosphate group by protein kinases to activate deactivated proteins.
Carboxylation
Addition of a carboxylic acid group, usually serves as calcium binding sites.
Glycosylation
Addition of oligosaccharides (carbohydrate) as proteins pass through the ER and Golgi apparatus to determine cellular destination.
Prenylation
Addition of lipid groups to certain membrane-bound enzymes.
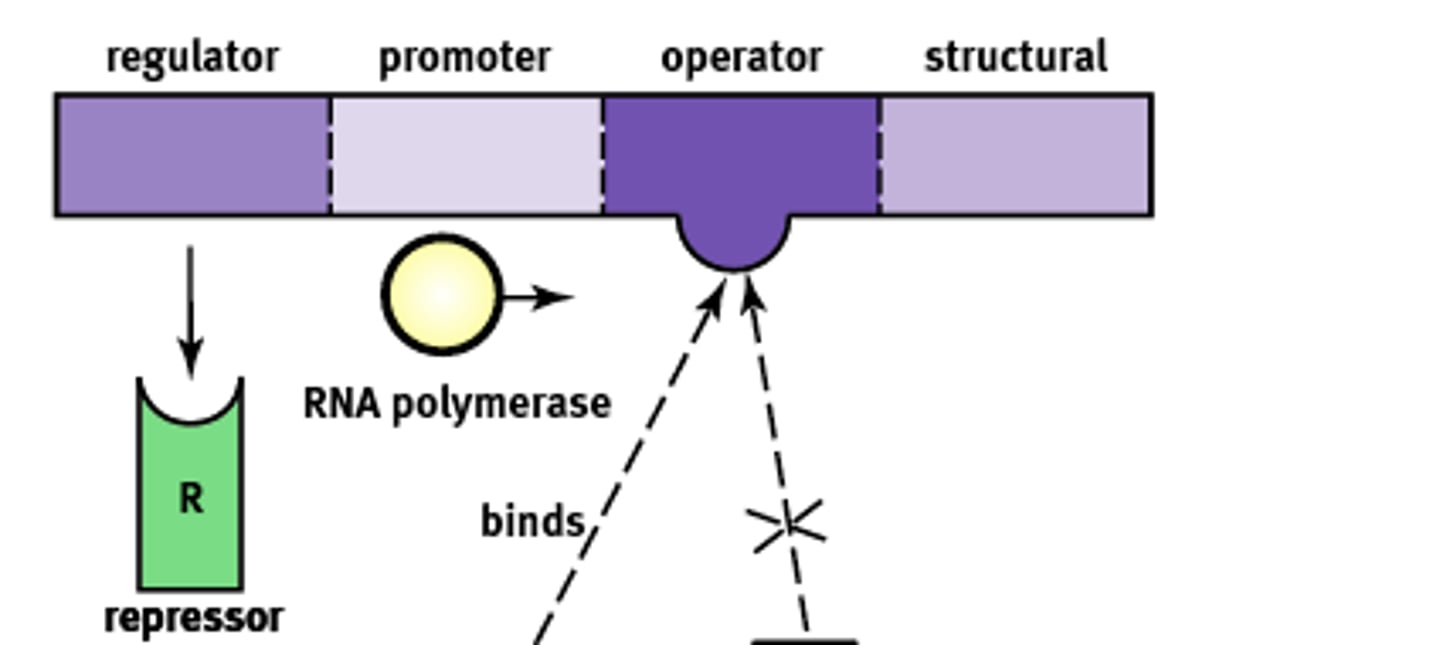
Operon Structure
Essentially an "on-off" switch that regulates gene expression. Composed of:
1) Regulator
2) Promoter
3) Operator
4) Structural
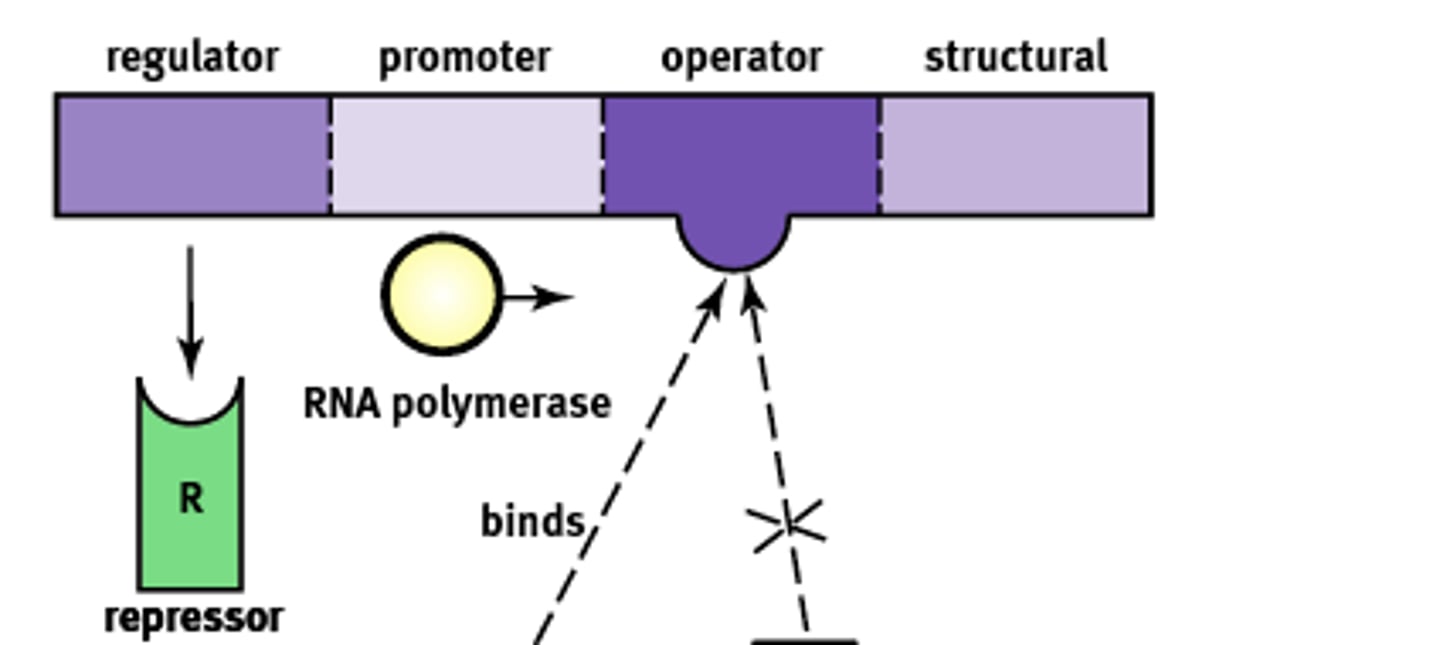
Regulator gene
Codes for the protein known as a repressor that prevents gene transcription.
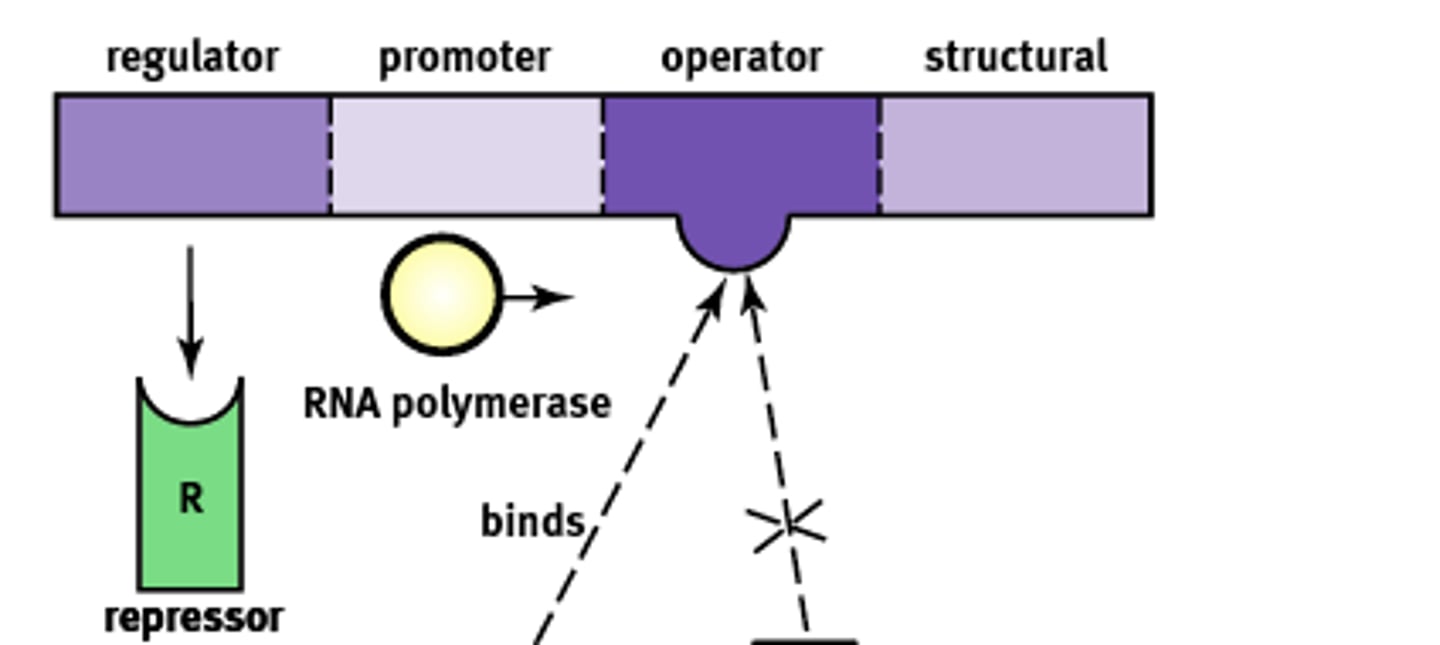
Promoter site
Site where RNA polymerase will bind to begin transcription. About ~25 base pairs upstream
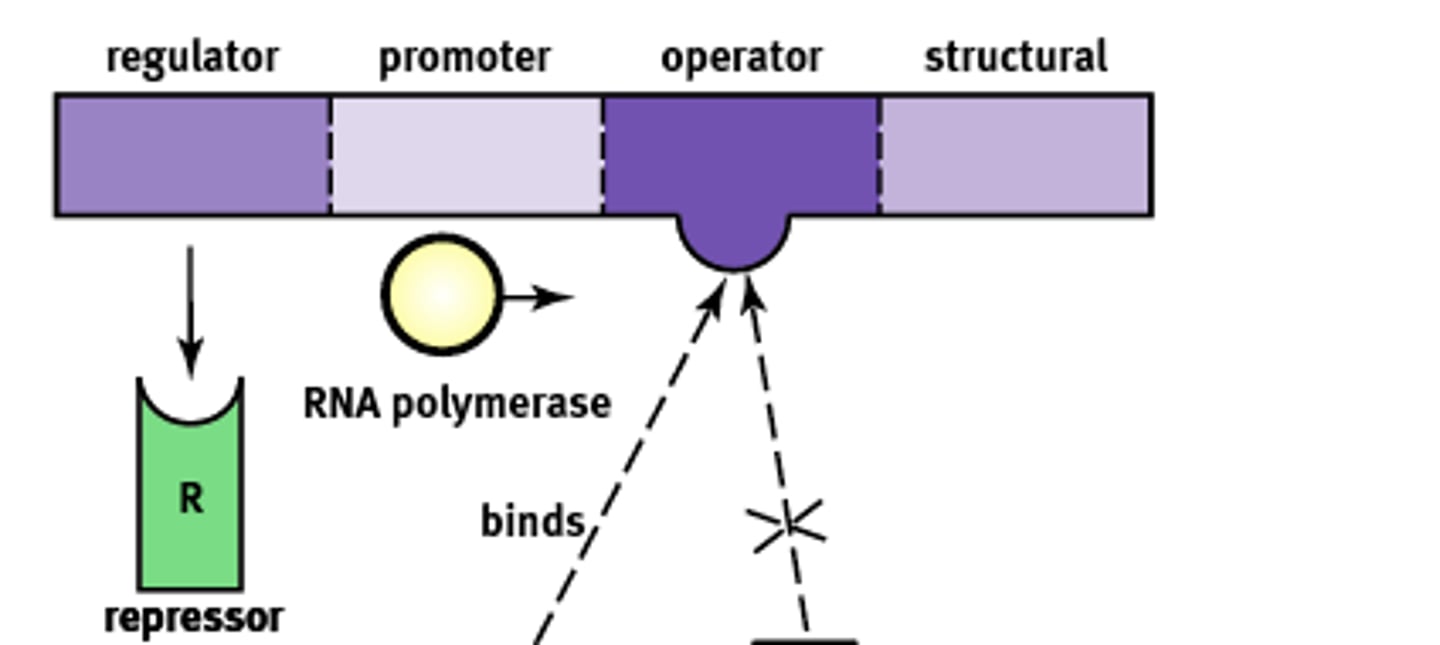
Operator
A non transcribable region of the DNA that is capable of binding a repressor protein and preventing transcription.
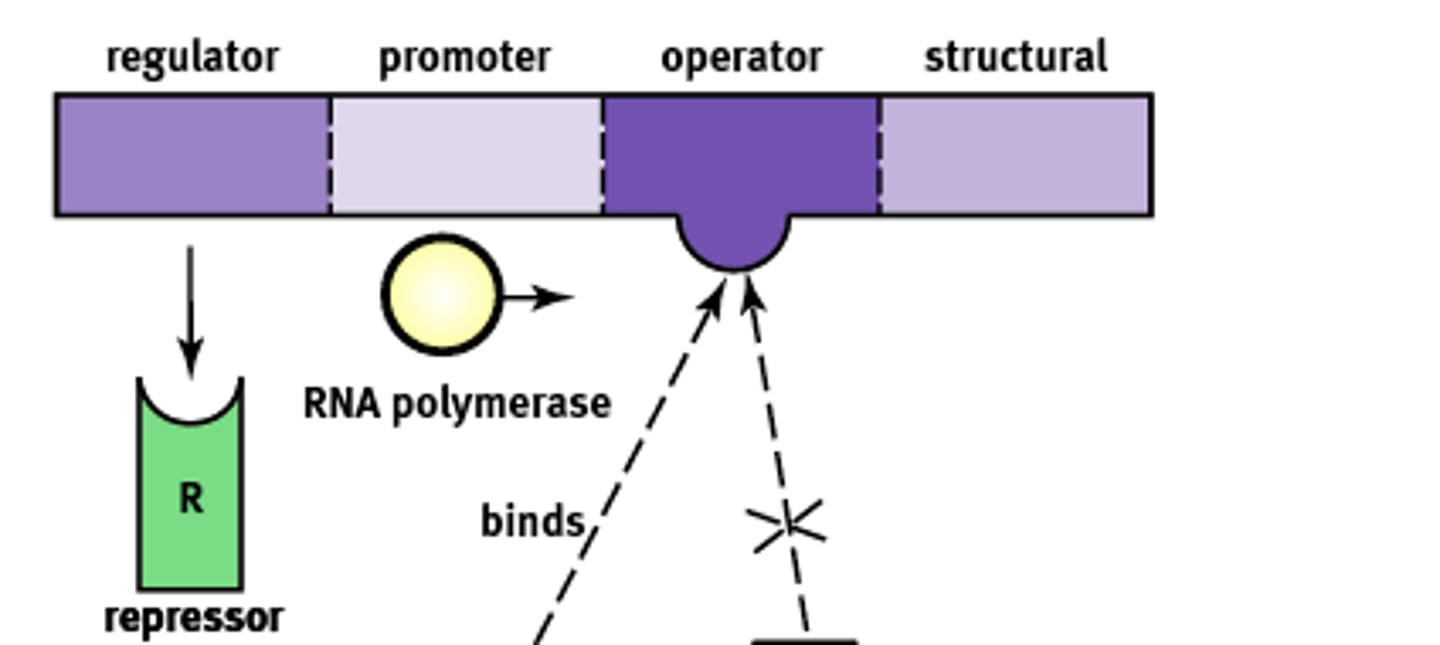
Structural gene
Codes for the protein of interest
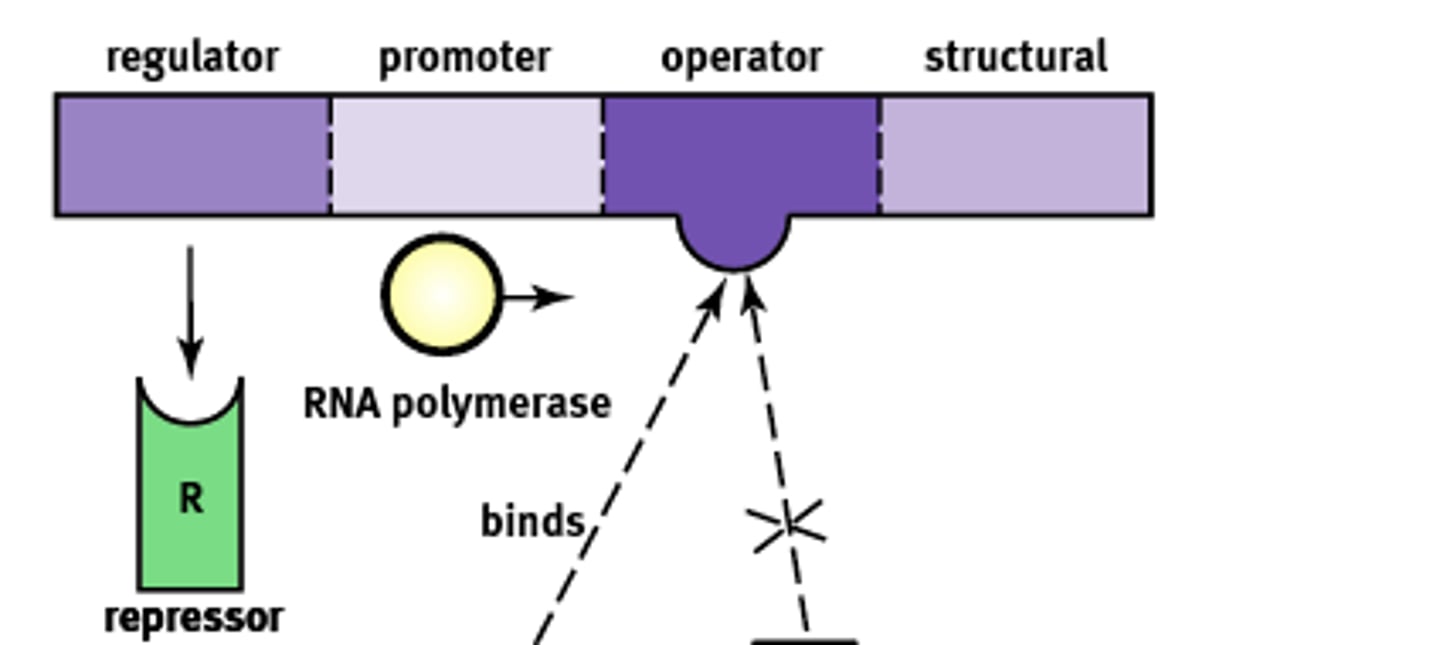
Inducible system
Systems that are bonded to a repressor under normal conditions. (usually off)
1) They can be turned on by an inducer pulling the repressor from the operator site.
Ex) lac operon.
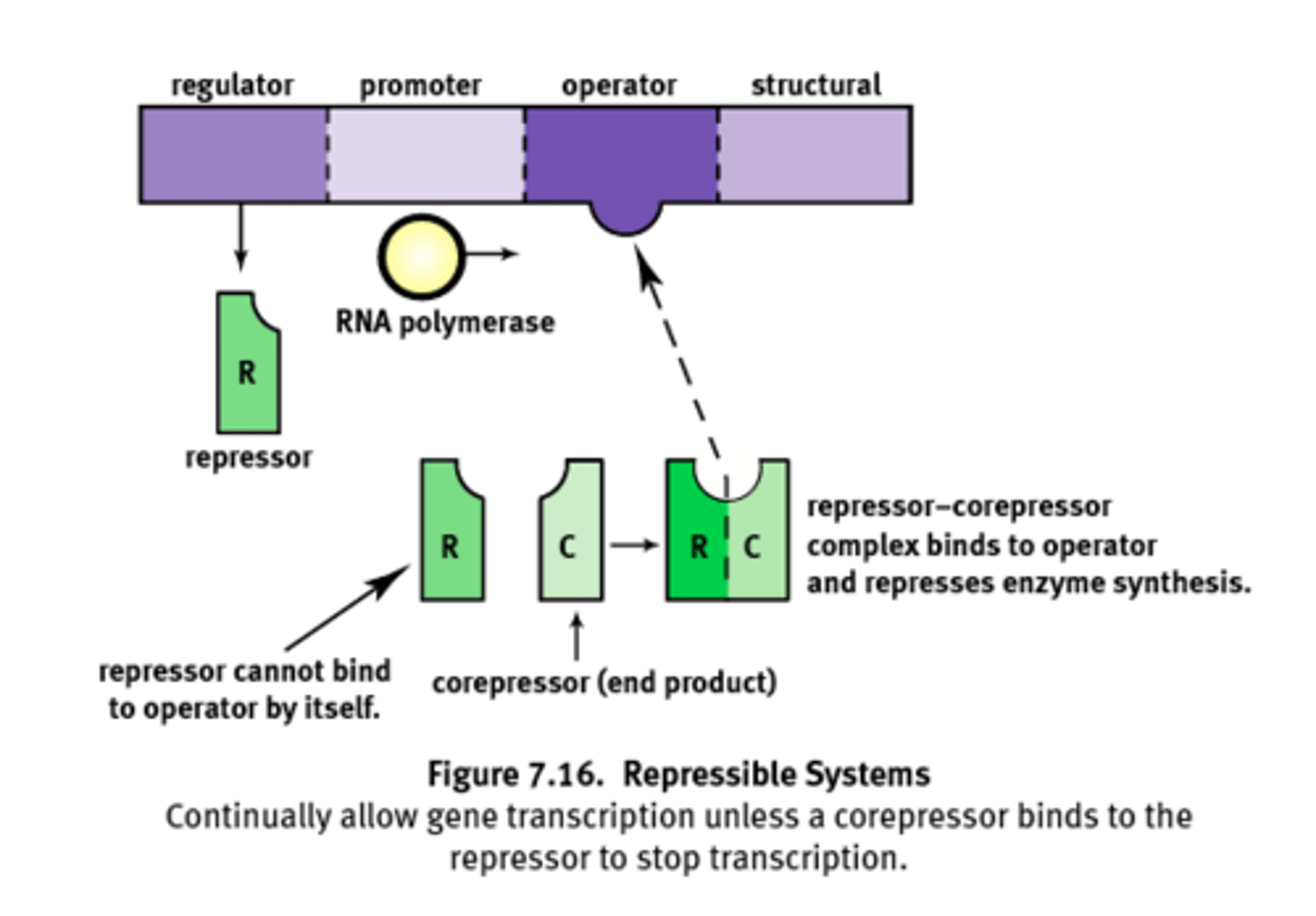
Repressible system
System that is transcribed under normal conditions. (usually on)
1) Can be turned off by a corepressor coupling with the repressor and binding to the operator.
Ex) Trp operon
Transcription factors
Collection of proteins that mediate the binding of RNA polymerase and the initiation of transcription.
-increase the rate of transcription
-2 domains: DNA binding domain and activation domain
Enhancers
Are more than 25 base pairs away from the transcription start site.
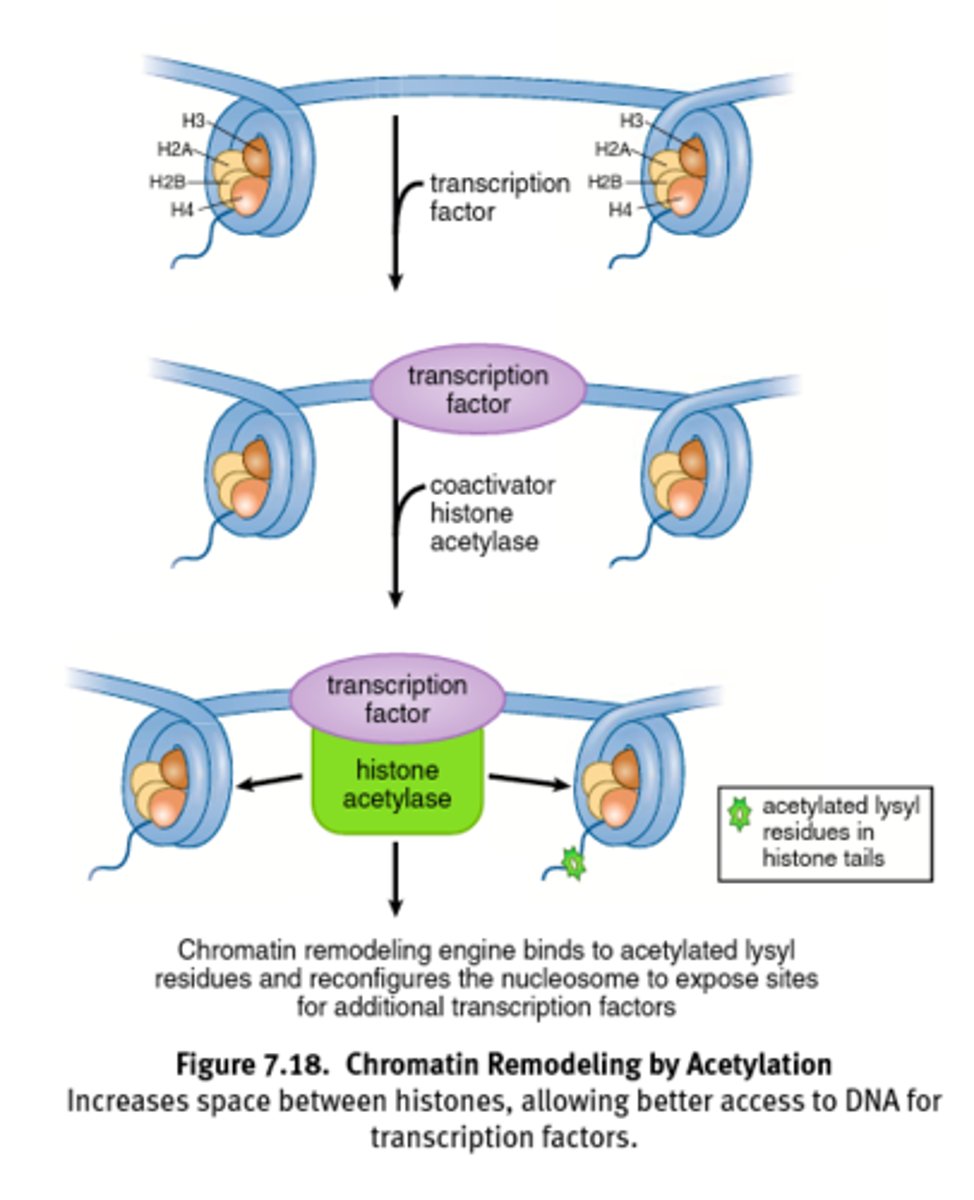
Histone Acetylation
Acetylation of histones decreases positive charge on lysine residues and weakens histone-DNA interactions.
Results in open euchromatin formation that allows for easier asses for transcription factors.
Acetylation = Access
DNA methylation
Add methyl groups to cytosine and adenine nucleotides. Methylation of genes is often linked with silencing gene expression.
The heavily methylated DNA hinders transcriptional machinery.
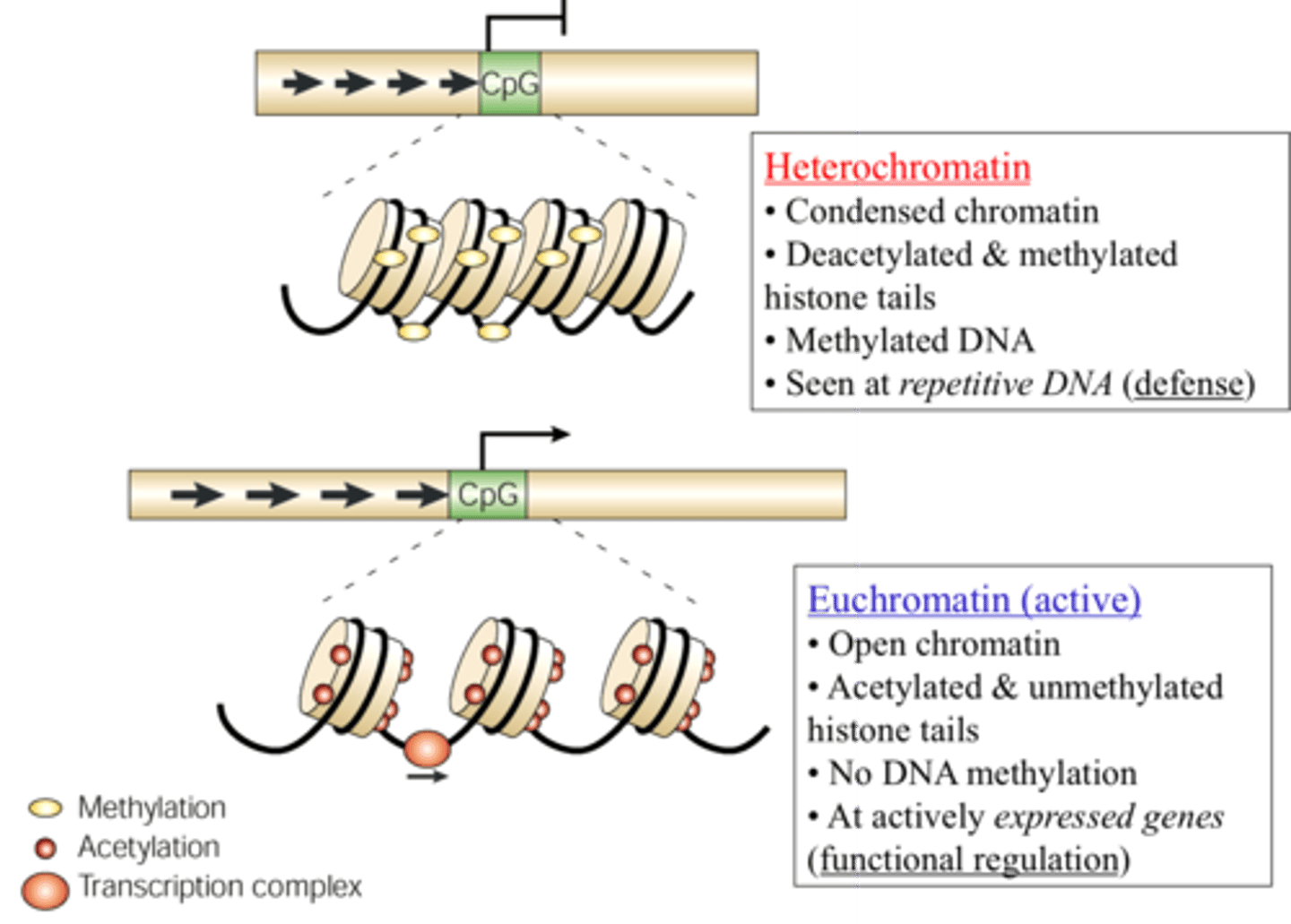
How to make DNA open/ closed for transcription?
Closed = methylated DNA that will pack tightly into heterochromatin.
Open = acetylation of histones causes them to form euchromatin that is open for business.
If a DNA sequence is 5' - TCTTTG - 3', what is the transcribed RNA strand?
5' - CAAAGA - 3' because mRNA is antiparallel to DNA