Histone Modification: DNA Methylation and Acetylation
1/32
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
33 Terms
RECAP: What is epigenetics?
A mechanism for stable maintenance of gene expression that involves marking DNA or its proteins
RECAP: What is the function of epigenetics?
To make genotypical identical cels be phenotypically distinct
RECAP: What are the other control mechanisms?
DNA methylation and chromatin modification
RECAP: What 3 processes control epigenetics?
RNA interference
DNA methylation
Histone modification
RECAP: How is DNA packaged in most cells?
Packaged with histone to from nucleosome in '“beads on a string” structure
RECAP: Chromatin regulation
What does it involve?
How can further regulation be achieved?
Which histones are prone to be post tanslational
Involves: High-order conformational changes
Such as relaxation or tightening of the thread of DNA-histone complex
Further regulation achieved by: Assembling promotor-enxchanes compaxes through long range DNA looping
Which can be blocked by specific DNA sequence
Histones: Subject to promoter-enhancer complex. Such as =
Methylation
Acetylation
Histone Modifications
What
Results
How can it change throughout lifespan
Role
What: Key epigenetic regulators that control chromatin structure and gene transcription
Results: Impacts phenotypes of similar genotypes
How can it change throughout lifespan: Because the abundance and localisation is responsive to environmental stimuli
Role:
DNA transcription
DNA repair
DNA replication
What are 2 diseases that is caused by insufficient histone modification?
Coffin-Lowry syndrome
Rubinstein
Coffin-Lowry Syndrome
Characteristics
Caused by
Inherited as
Males or females suffer more?
Characteristics:
Mental retardation
Abnormalities of head and face
Caused by: Mutation in the RSK 2 gene (histone phosphorylation)
Inherited as: X linked dominant
Males or females suffer more: Males
Rubinestein-Taybi Syndrome
Characteristics
Caused by
Characteristics:
Short
Intellectual disapbility
Distinctive factial features, board thumbs and free toes
Caused by: Mutations in CREB-biniding protein (histone actylation)
What is DNA methylation?
The addition of methyl group to C-5 position of cytosine residue at CpG dinucleotides
*Human genome is not methylated uniformly, containing regions of methylated and unmethylated
In what kind of organisms does DNA methylation occur in?
Occur in cells of fungi, plants, non-vertebrates and vertebrates
What is the percentage of methylated DNA cytosine in vertebrates and plants?
Vertebrates = 3-6%
Plants = 30%
In what organism does DNA methylation DOES NOT occur?
Many insects and single-celled eukaryotes
What is the function of DNA methylation?
Turns off genes
Inhibit transcription
What is DNA methylation a therapeutic target?
The change from DNA methylation is heritable and reversible
When is DNA methylation determined?
During embryogenesis and is passed over to differentiating cells and tissue
Where does methylation occur?
Occur at:
In cytosine in sequence context 5’CG3’ that is immediately followed by guanine-CpG dinucleotide
Non-coding regions
Interspersed repetitive elements
DOESN’t occur at:
CpG islands of active genes
When does methylation typically occur?
After DNA replication
What is the mechanism?
Methyl groups are transferred from S-adenosyl methionine (SAM)
In a reaction catalysed by a DMNT
Causes SAM to be converted into SAH (S-adenosyl homocysteine)
What are the 3 types of methylation processes
Maintenance methylation: Refers to the preservation of existing DNA methylation patterns after DNA replication
Requires a hemi-methylated DNA substrate
De Novo methylation: Refers to the addition of methyl groups to unmethylated DNA sequences, producing new methylation patterns
Demethylation: Refers to the removal of methyl groups from DNA
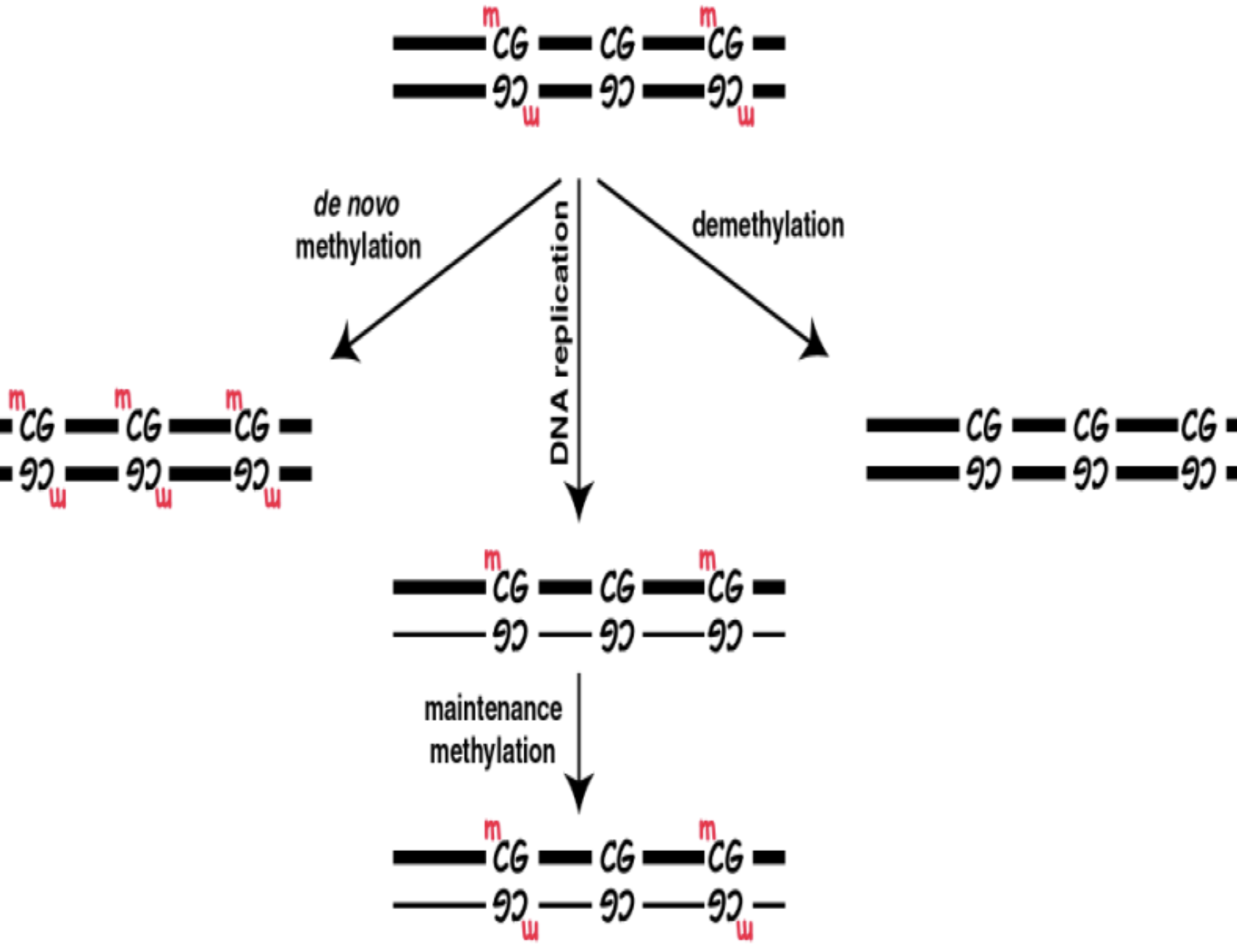
Control of DNA methylation
By what
What are the 5 classes of them?
Which 3 is the most important and their function
What controls DNA methylation: DNA methyltransferase (DNMTs)
5 classes:
DNMT1
DMNT2
DMNT 3a and DMNT3b
DMNT3L
Most important 3 and their functions:
DNMT1: For maintenance methylase
DMNT 3a and DMNT3b: For de novo DNA methylation
What kind of methylation process would occur in these situations =
Pre-implantation
After implantation of embryo and during carcinogenesis
During replication
Pre-implantation: Genome undergoes demethylation
After implantation of embryo and during carcinogenesis: Novo methylation creates new methylation patterns
By DMNT 3a and DMNT 3b
During replication: Maintenance methylation allows methylation patterns to be maintained
DMNT1 methylates the hemimethylated DNA after strand synthesis
What are the 3 types of methylated bases in DNA?
C5-methylcytosine(5-mc)
N4-methylcytosine
N6-methyladenine
What are the 5 effects of DNA methylation?
Deactivaton of parasitic Transposons
Somatic hyper-mutations at lg locus in B and T cells
Embryonic development and growth
Genomic imprinting
X-chromosome inactivation
Heterochromatin maintenance
Tissue specific expression controls
Silencing of repetitive elements
CpG islands
What
Methylated or unmethylated
Relationship between non-methylated regions and transcription
What occurs at promotor when methylation occurs
What occurs at non-promotor when methylation occurs
Amount in human genes
Present in what type of gene
What: Regions with high concentration of phosphate-linked cytosine-guanine pairs (cytosine nucleotide is followed by guanine nucleotide)
It can be: Both unmethylated or methylated
Unmethylated at promotor regions
Methylated at non-promotor regions
Correlates to transcriptional silencing due to the inhibition of transcription factor binding
Relationship between non-methylated regions and transcription:
Allows binding of transcription factors
Allowing transcription to occur
HOWEVER there are cases where even though CpG island is unmethylated, the gene is still silent
What occurs at PROMOTOR when methylation occurs:
Inhibition of TF binding
Causes transcriptional silencing
What occurs at NON-PROMOTOR when methylation occurs
Silence parasitic genetic elements
Genomic stability
Amount in human genes: Half of all genes have CpG islands
Amount in genome: Low abundance
Present in:
Housekeeping genes
Genes with tissue-specific patterns of expression
Dysregulation of methylation process results in?
Many disorders like:
Cancers (Deactivation of tumour suppressor genes
Imprinting disorder:
BWS (Beckwith-Wiedemann syndrome)
PWS (Prader-Willi syndrome)
TNDM (Transient neonatal diabetes melllitus)
Repeat-instability diseases
FRAXA (Fragile X syndrome)
Facioscapulohumeral muscular dystroph
Defects of the methylation machinery
SLE (Systemic lupus erythemtosus)
ICF (Immunodeficiency, centromeric instability and facial abnormalities)
How can methylation imbalance contribute to tumor progression?
Global HYPOmethylation
Activates oncogenes and genomic instability
DNA HYPERmethylation
Inactivation of tumor suppressor genes
Inactivation of DNA repair genes
What can cause DNA hypermethylation? (risk factors)
Carcinogenes
Because chronic exposure of human bronchial epithelial cells to tobacco-detrived carcinogens drives cells to tobacco-derived carcinogens
What can cause DNA HYPOmethylation? (risk factors)
Cigarette smoke
What are other 2 risk factors?
Reactive oxygen species (ROS)
Aging
What are the 5 ways DNA methylation can be detected?
Sodium bisulfite conversion (SBC)
SBC LC-MS-MS
cDNA microarray
Restriction landmark genomic sequencing
CpG island microarray
Sodium bisulfite conversion (SBC)
Allows the highest degree of resolution of methylation status
Therefore what does it determine?
Involves?
How does it detect methylated regions?
What is done after
Therefore what does it determine: The positional CpG genotype
Involves: The chemical modification of DNA by bisulfite treatment
Sodium bisulfite converts cytosine to uracil
How does it detect methylated regions: Methylated cytosine is resistant to this conversion
What is done after:
PCR to amplify alleles
Methylation-specific polymerase chain reaction is most useful