Transcription and Translation (Midterm 3 Lecture 2)
1/49
Earn XP
Description and Tags
51 slides
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
50 Terms
what synthesizes rna transcripts, and which rna is the rna transcript in prokaryotes and eukaryotes
rna pol transcribes dna to produce rna transcript
pro = mRNA
euk = pre-mRNA (further processed to become mRNA)
what translates mRNA sequence to synthesize a polypeptide
ribosomes and tRNA (transfer rna)
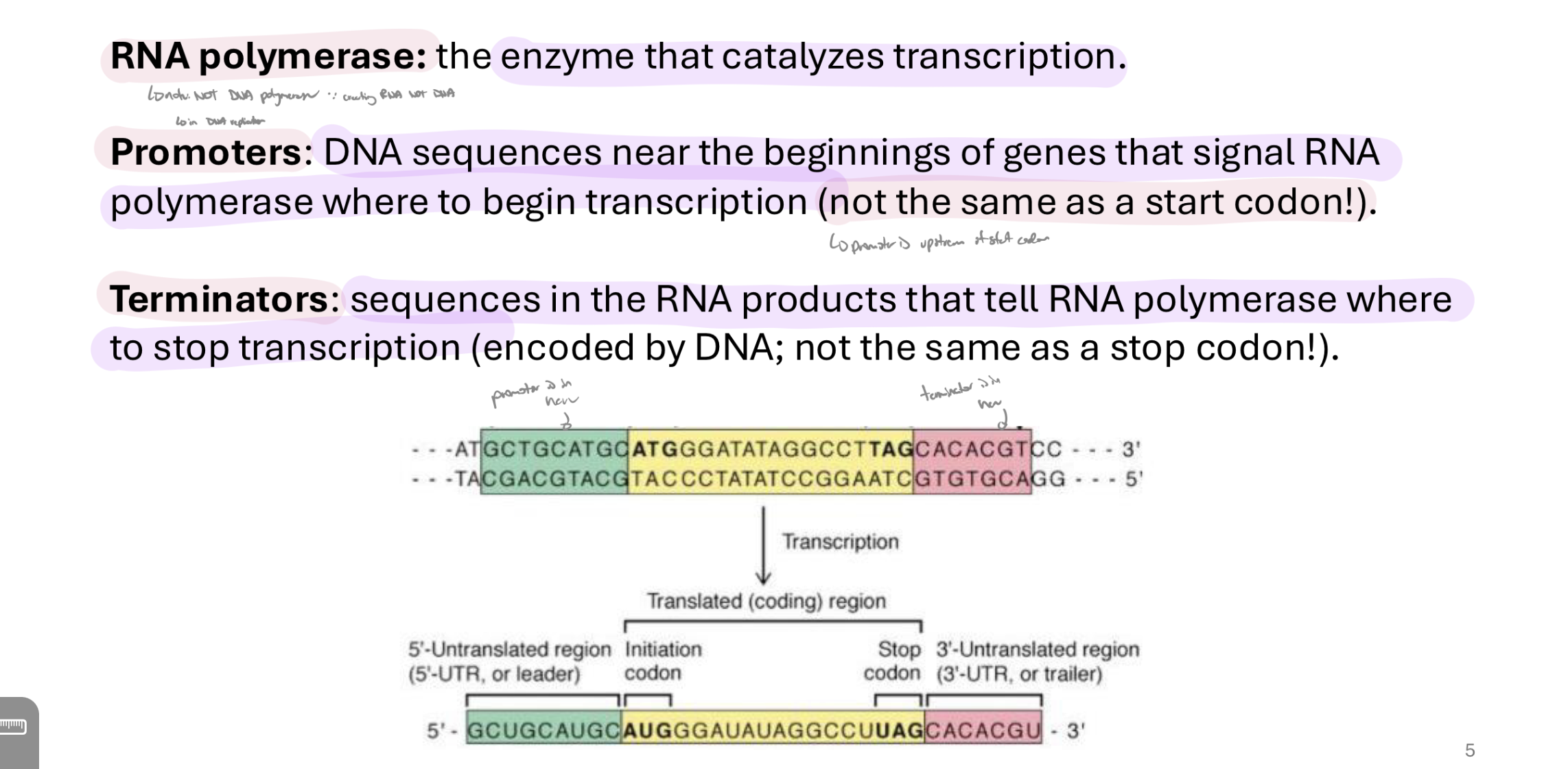
what is rna polymerase, promoters, and terminators
notes:
rna pol is diff from dna pol → rna pol transcribes, dna pol dubpicates dna
promoters are NOT the same as a start codon → they are upstream from them
same w terminators and stop codons → terminators are downstream
explain the initiation phase of transcription in prokaryotes
rna pol binds to promoter sequence
sigma factors bind to rna pol to form the holoenzyme
region of dna is unwound to form an open promoter complex
unwinding lets phosphodiester bonds form btwn first 2 ribonucleotides
no primer needed
only template, can start new rna molecule from scratch
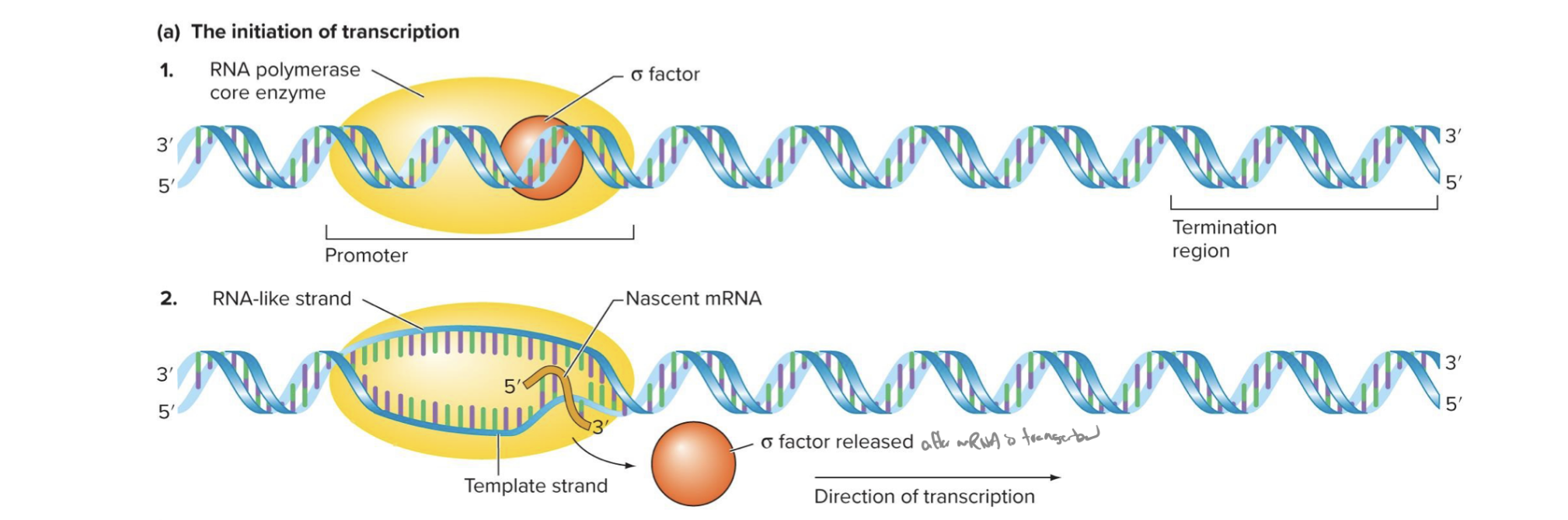
what is a holoenzyme composed of
sigma factors + rna pol
what does nascent mean
coming into or “future”
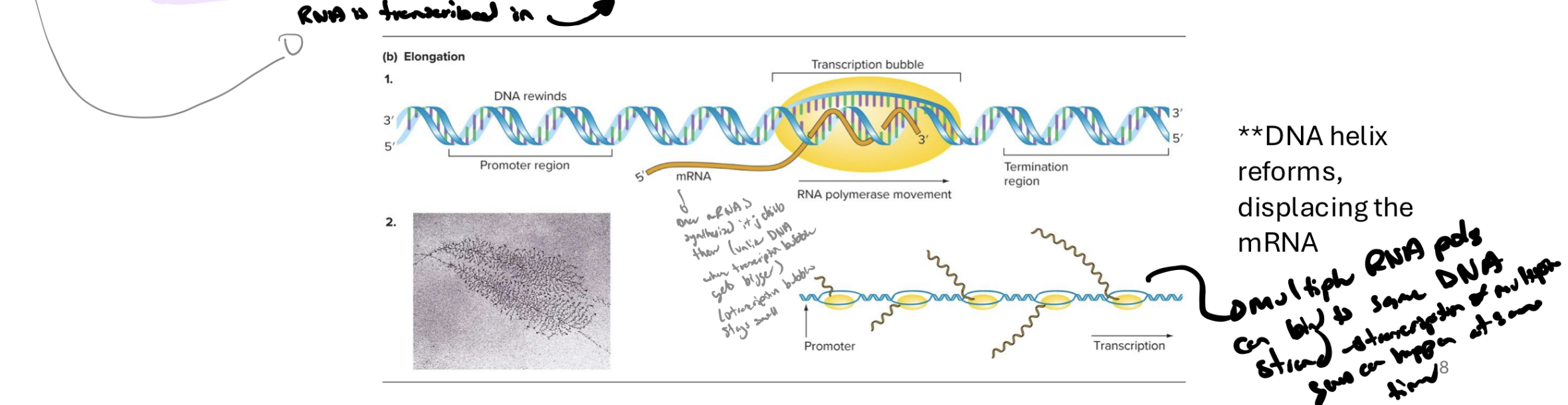
explain the elongation phase of transcription in prokaryotes
sigma factor is released from rna pol (rna pol is now the core enzyme)
rna pol loses affinity for promoter and moves along template strand (in 3’ to 5’ to synthesize new rna in 5’ to 3’)
NTPs are added to the nascent (fture) mRNA
complimentary base pairing w dna
uracil is incorporated insted of thymine (both pair w A)
nucs are added in the 5’ to 3’ direction
why do framshifts in one gene not affect frameshifts in other genes in translation/transcription
bc whole there are non-transcribed (spacer) regions of dna → so each gene is transcribed separately
explain the termination phase of transcription in prokaryotes
transcription stops when terminator sequence is transcribed
what are the 2 kinds of terminators in prokaryotes? explain them
extrinsic: require rho factor
rho protein binds to rna sqeunece that is C-rich and G-poor without secondary structure (on its own, no dna or anything bound to it (bc rna is single-stranded))
intrinsic: (dont require additional factors)
no protein involved
GC-rich rna region makes a hairpin structure when mRNA is transcribed → the stucture pulls rna away from polymerase
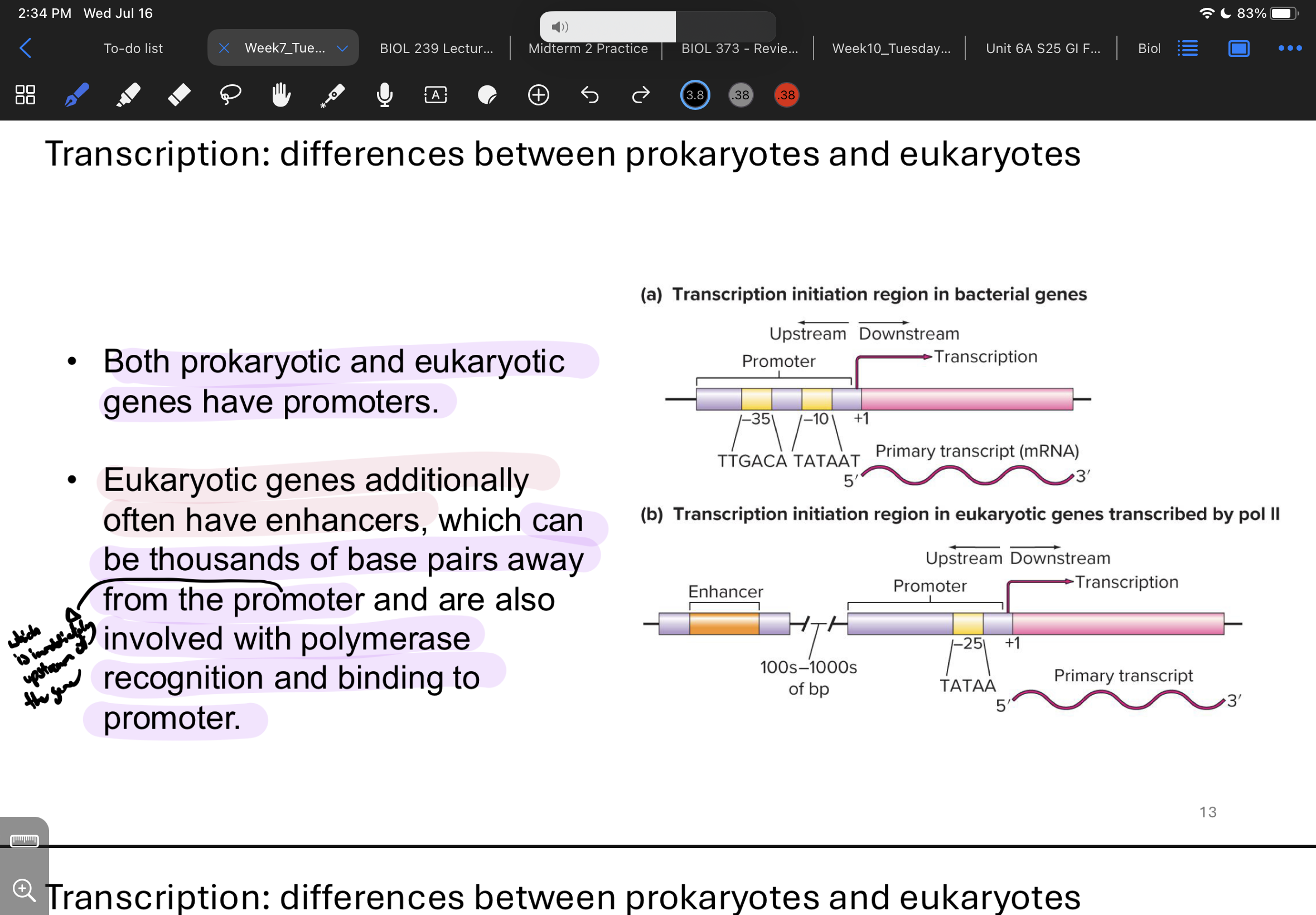
what are some diffs between transcription in prokaryotes vs eukaryotes
both have promoters BUT only eukaryotic have enhancers (which can be thousands of bps away from promoter (which is immediately upstream of the gene)
t/f: mRNA and protein sequences can always be directly deduced from the dna sequence
false → only in prokaryotes (“what you see is what you get” → euks have additional stages of processing after initial transcription
what are introns and which types of organisms have them
segments of dna that are transcribed but not translated
ONLY in euk genes (not proks)
what are some modifications of primary rna transcript to produce mRNA in euks
rna is spliced to remove introns (non-coding regions of dna)
5’ methyl cap is added
poly-a-tail is added
how is the 5’ methyl cap added on eukaryotic mRNA made and what does it do
capping enzyme → adds guanidine triphosphate (the nuc) in reverse oriegntation to the 5’ end after polymerization of the transcript’s first few nucleotides
that guanine is NOT encoded for by the gene
methyl transferases then add methyl groups to the backwards G and one or more of the succeeding nucs in the RNA
it is critical for efficient translation, transportation out of nucleus, and preventing degredation by exonuclease (things that break down nucleic acids)

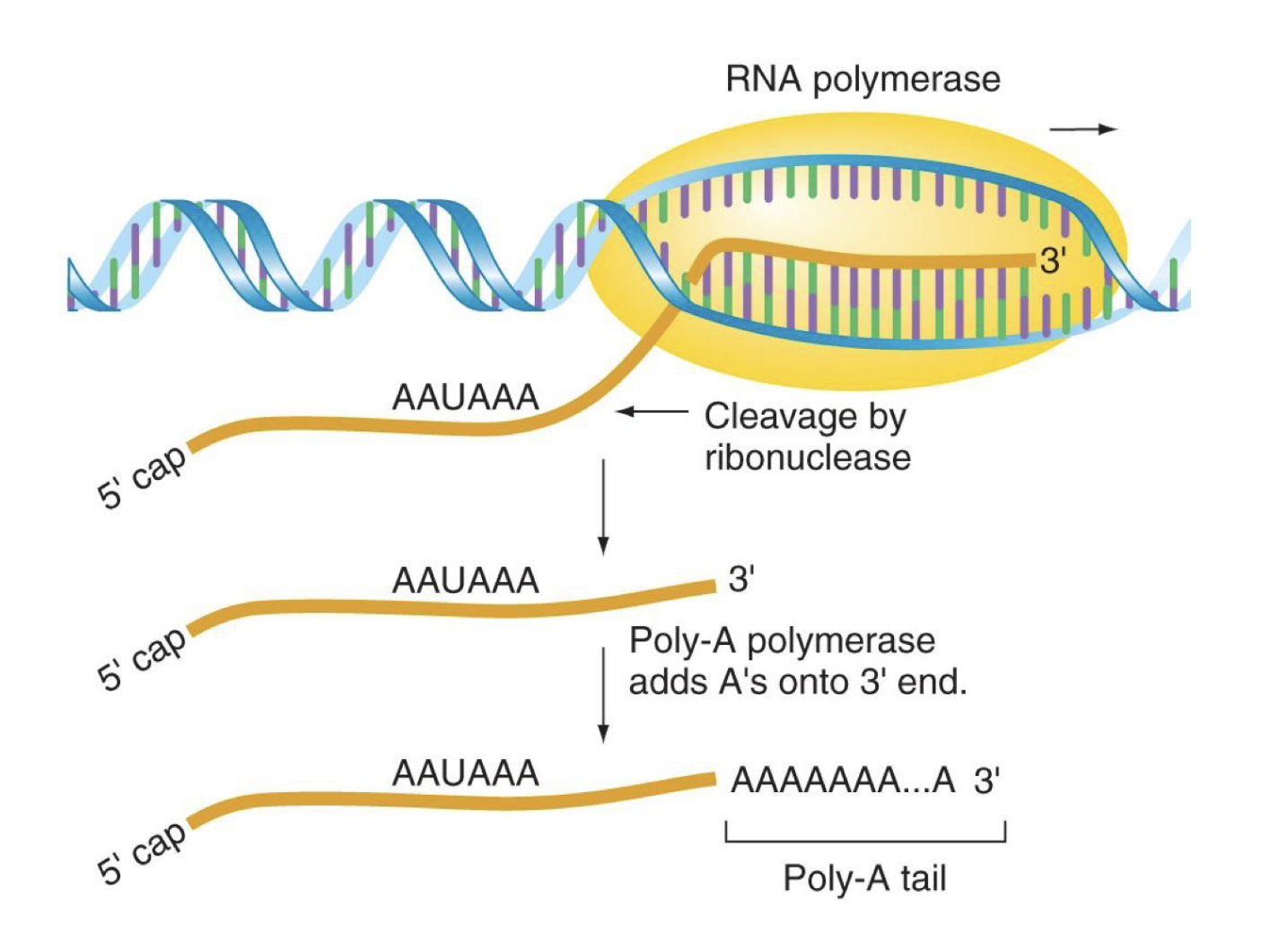
how is the poly-a-tail added on eukaryotic mRNA made and what does it do
adition of 100-200 adenosines to the 3’ end of the RNA
NOT encoded for by the gene
located 11-30 nucs downstream of an AAUAAA sequence
ribonuclease (RNase → cuts rna) cleaves primary transcript at terminator sequence then poly-a polymerase adds As to the 3’ end
critical for efficient translation, transportation out of nucleus, and preventing degradation by exonuclease (similar fx to 5’ methyl cap)
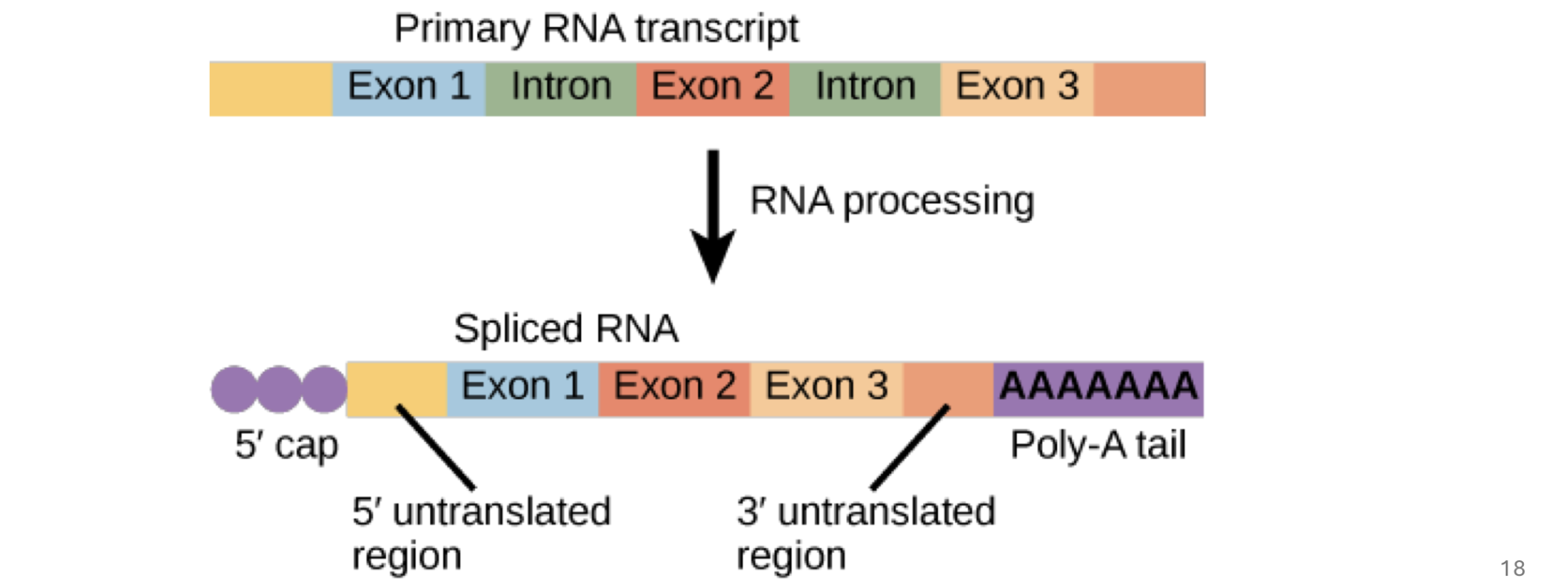
what are exons
sequences found in both a gene’s dna and the mature mRNA
exons contain coding sequences for protein product and 5’ and 3’ UTRs (untranslated regions)
what are introns
sequences dfound in a gene’s dna but NOT in the mature mRNA → they’re removed from the primary transcript through splicing
how are introns removed
through 2 sequential cuts
one end is cut by splicosome (usually, sometimes rna transcripts are self-splicing)
intron folds in on the other end of itself
the other end us cut
the mRNA fuses back tg and the spliced intron is degraded
do al euk genes include introns
no → some include multiple, some have none
what is the dystrophin gene? explain how it relates to splicing
the longest known human gene → goes from 25,000,000 bps to 14,000 after pslicing (extreme ex, usually not THIS much)
the human genome contains around 28,000 genes but human cells produce hundreds of thousands of diff proteins → how
alternative splicing
what does alternative splicing do
produces diff mature mRNA molecules from one gene
may encode related proteins w diff partially overlapping sequences
all exons of a gene can be transcribed, or a couple predetermined ones → hence how one gene can encode for multiple proteins w similar overlapping sequences

what is translation and what does it require to happen (what are the necessary components)
it is the process in which the genetic code carried by mRNA directs the synthesis of proteins
requires…
mRNA, tRNA (w attached AA), and ribosomes
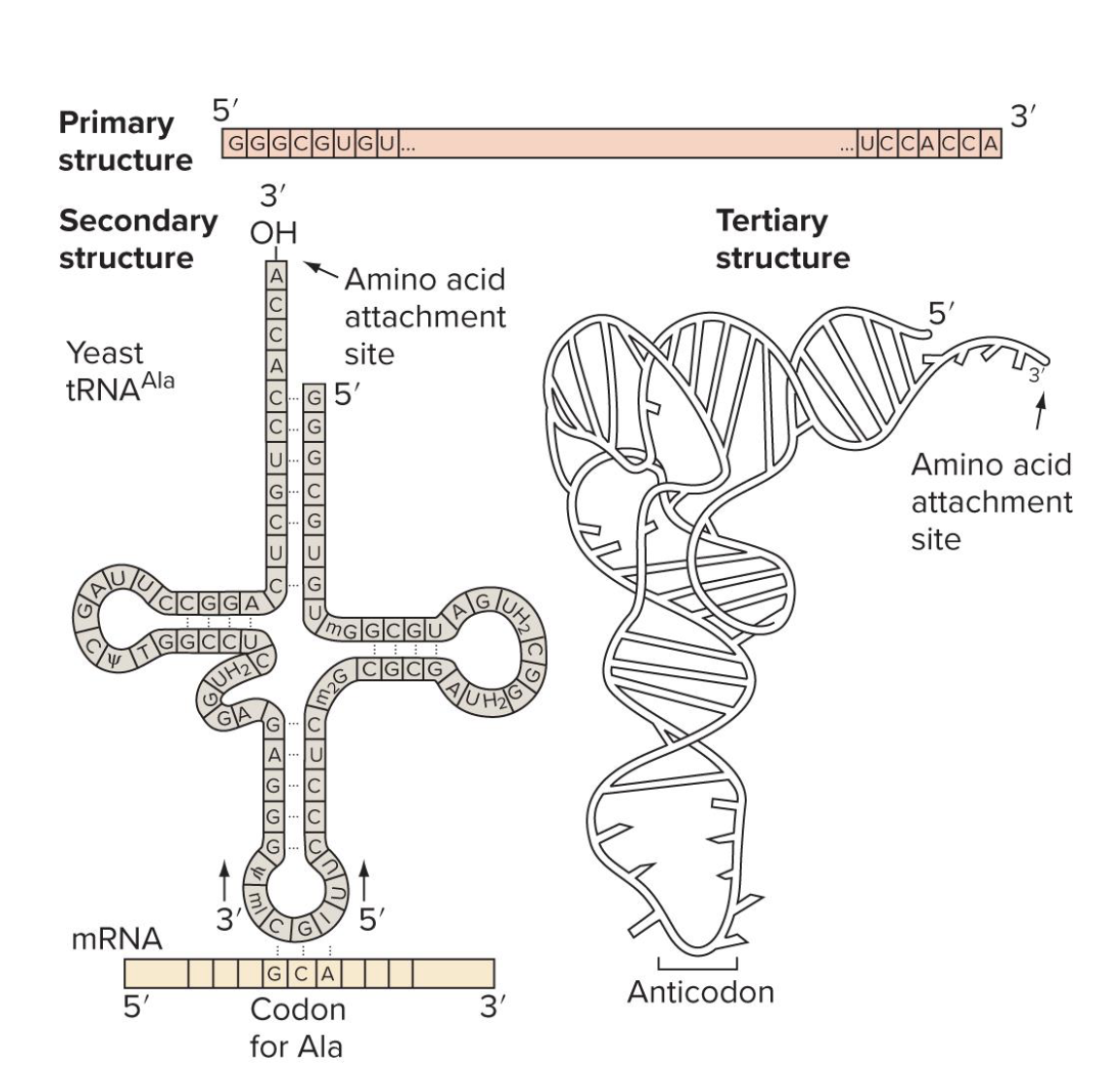
what is tRNA
short, single-stranded RNA molecules
each tRNA is covalently bound to a specific AA → (when that happens its referred to as “charged” tRNA)
base pairing btwn tRNA anticodon (complimentary to mRNA codon) and mRNA codon determine where an AA becomes incorporated into a growing peptide
explain the primary, secondary, and tertiary structure of tRNAs
primary: nuc sequence
secondary: base pairing of complimentary sequences within the tRNA (creates cloverleaf shape)
teritary: formed by 3d folding (L shape)
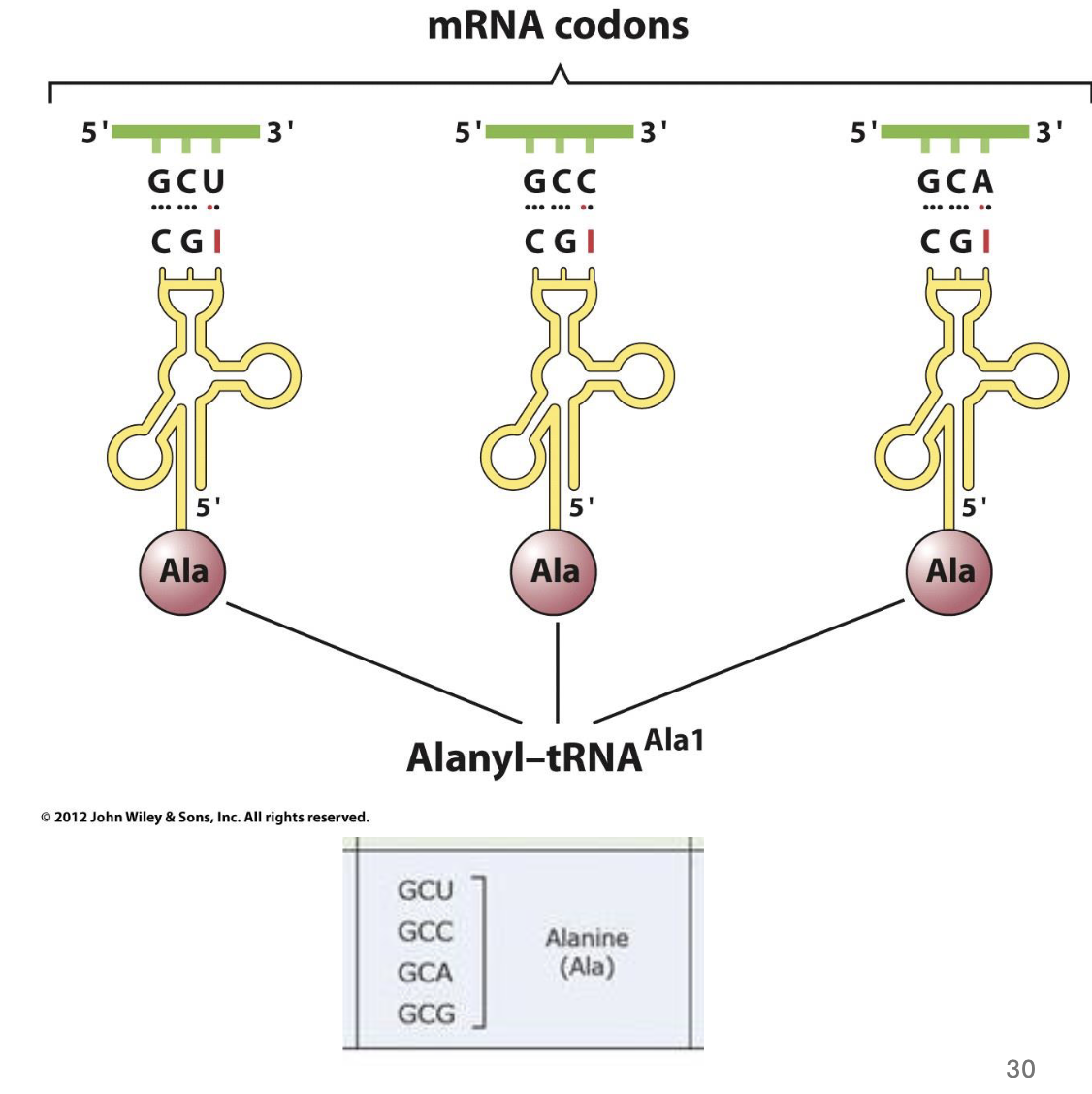
how many different codons are there for AAs
61 (plus 3 stop codons) → but there are fewer than 61 unique tRNA genes
what is wobble in translation
some tRNAs recognize more than one codon
the 3’ nucleotide in the codon often doesn’t add specificity to the codon
the nuc in the wobble position of the tRNA (the last one in the codon, 5’ end of tRNA molecule) is often modified
modified A,U,G, or C (w diff groups and stuff on them) → so it can bind to other things too since the last one usually doesn’t make a diff
which end of the tRNA molecule is the “wobble position”
the 5’ end of the tRNA molecule (bonds to the 3’ end of the RNA sequence)
note: wobble rules will be provided on exam
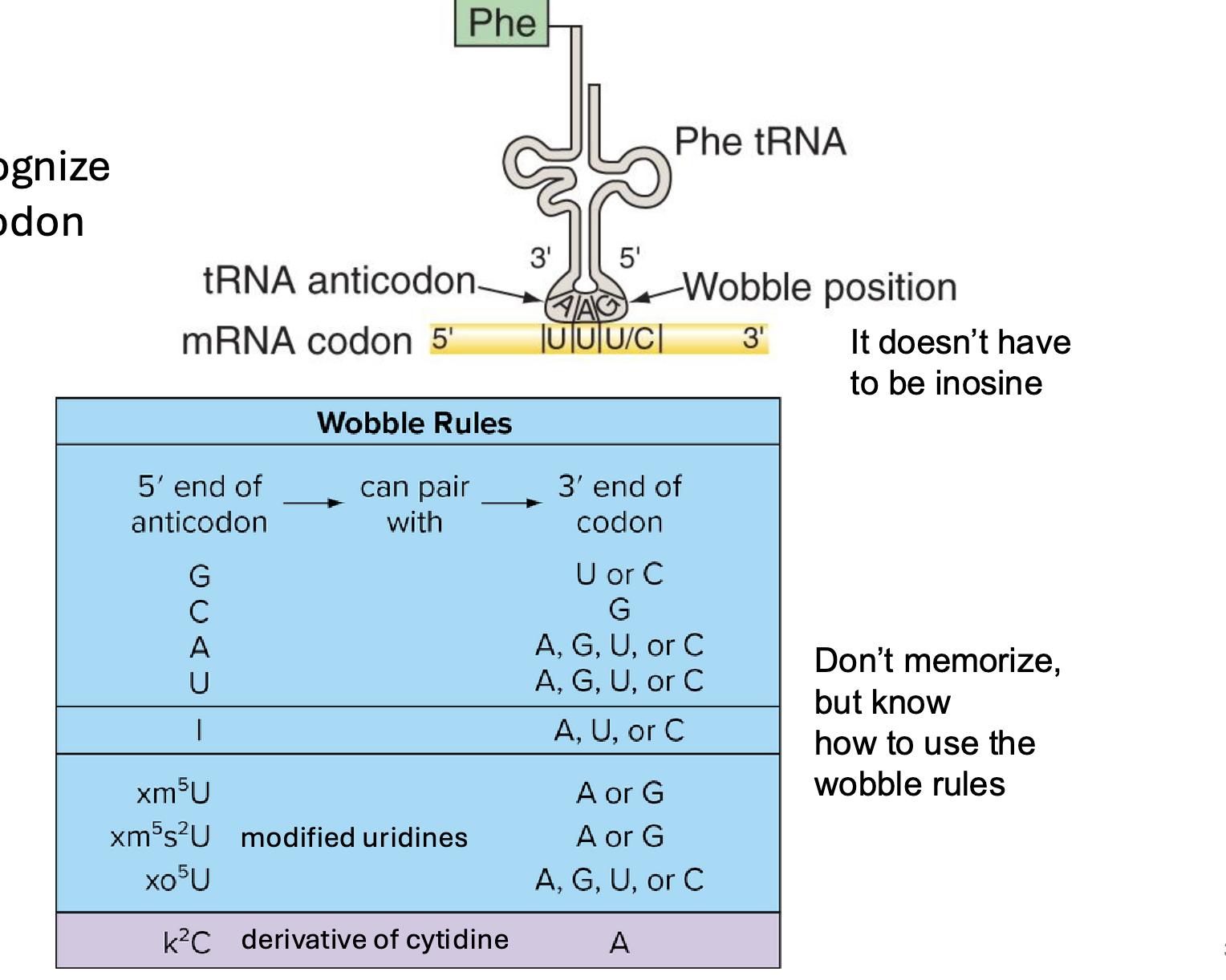
what is the I on the wobble rule chart
Inosine (a modified adenosine molecule)
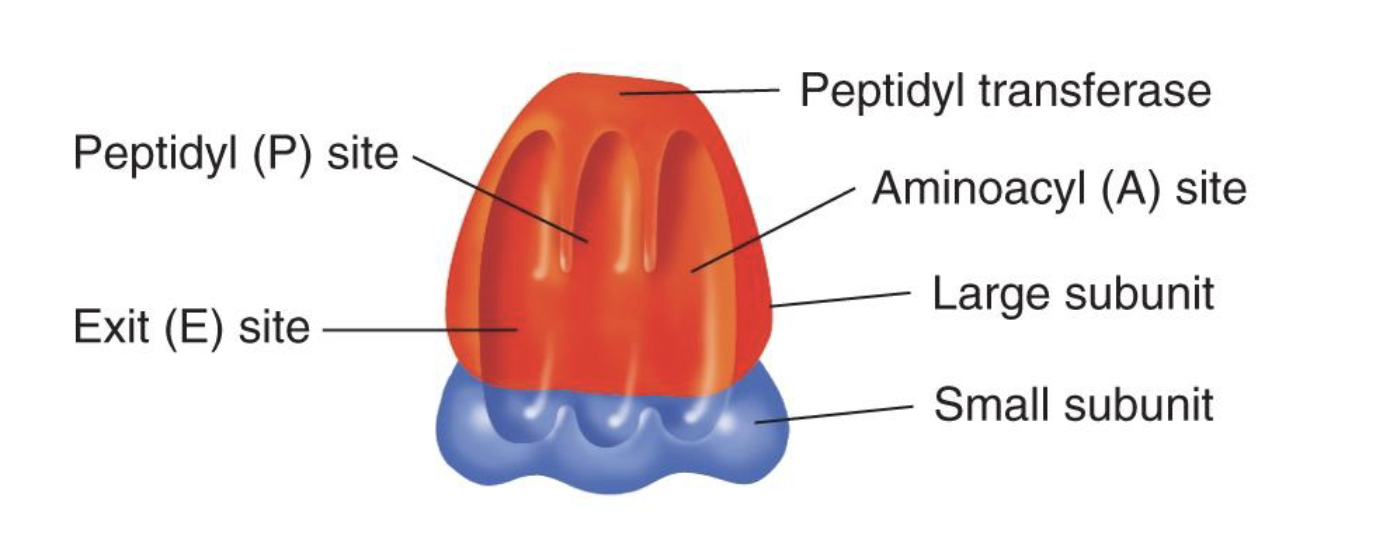
what is the ribosome and what does it do
the site of protein synthesis
composed of protein and RNA
has a large and small subunit
recognizes mRNA features that signal the start of translation
coordinates movement of tRNAs to match codons in mRNA
supplies enzymatic activity to link AAs and move 5’ to 3’ along the mRNA (backwards to way nucleic acids have been read so far)
helps facilitate termination by dissociating from mRNA and the polypeptide
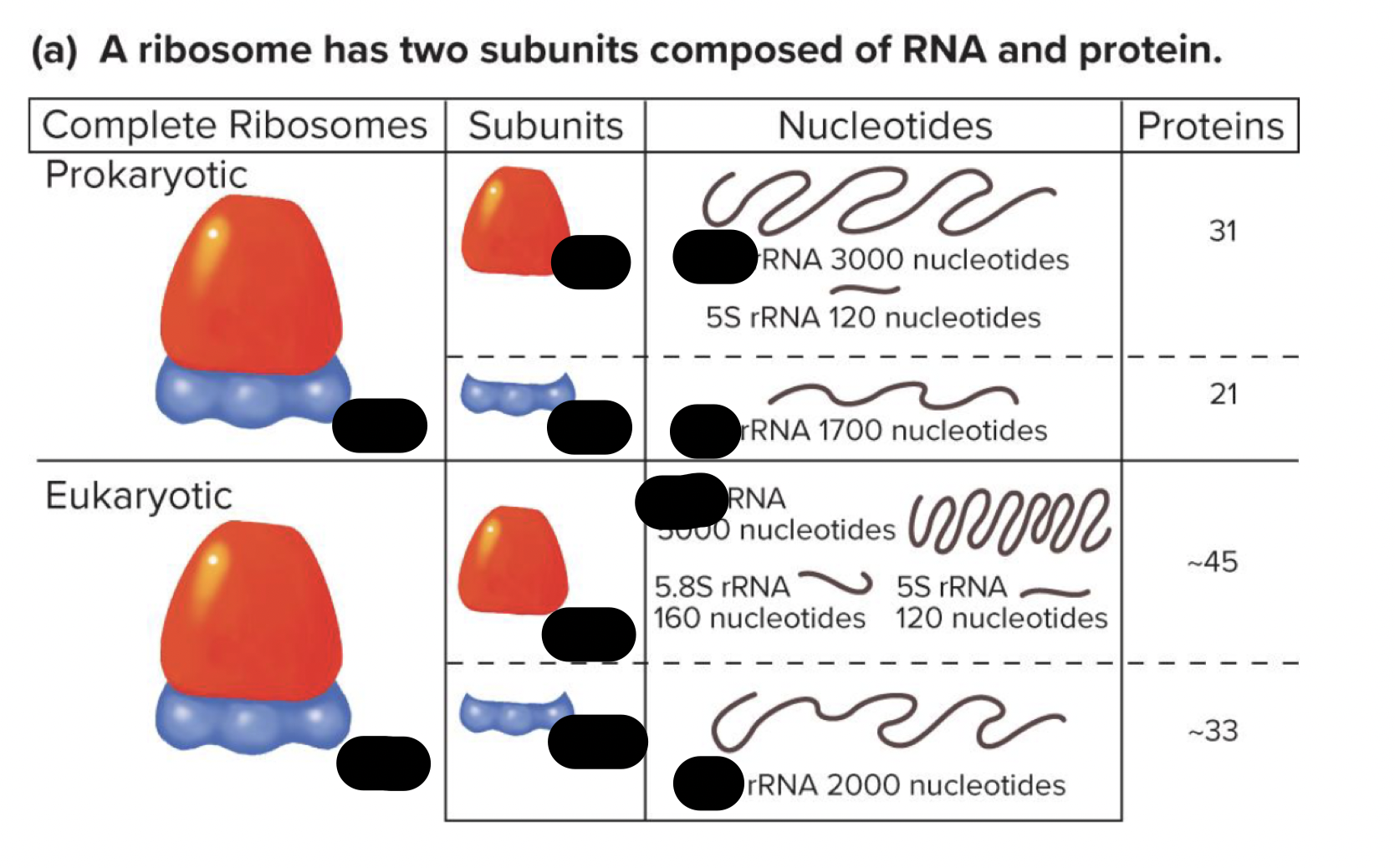
t/f: the sizes of ribosomal subunits differ bewteen pro and euks. explain why or y not (how big each subunit in both organism are)
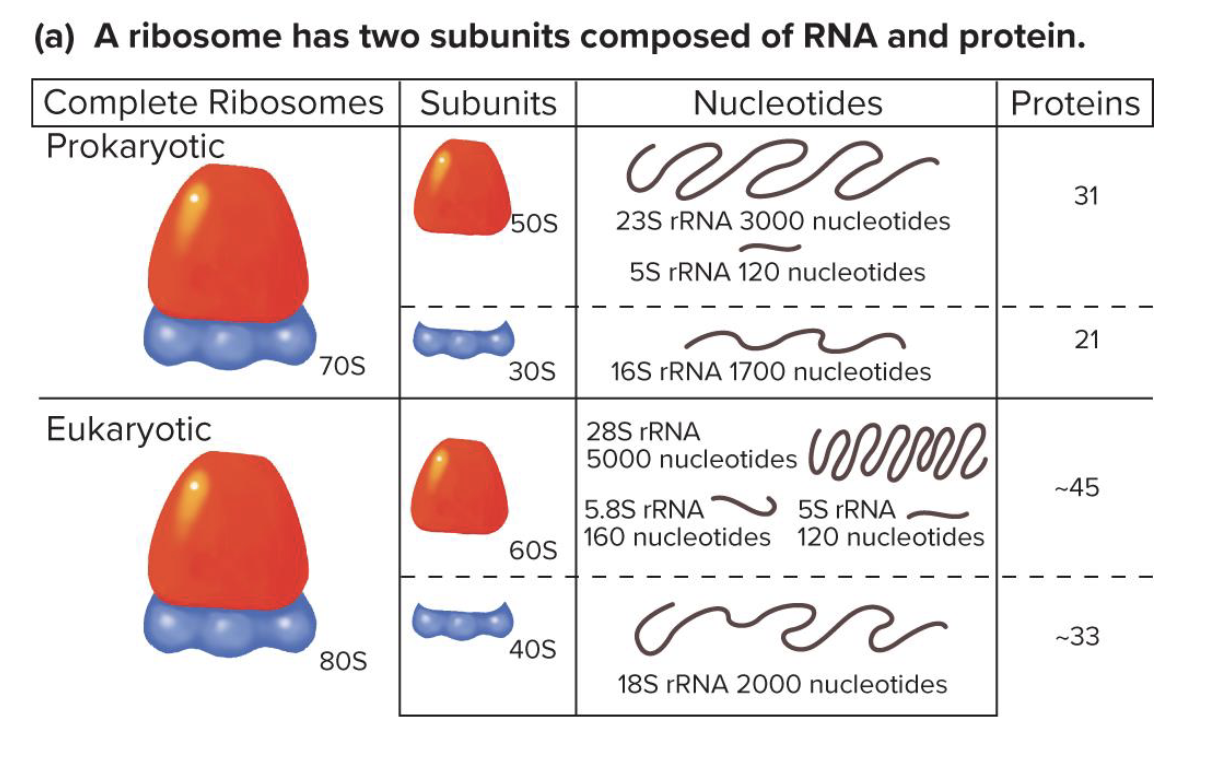
what do the small and large subunits of ribosomes do respectively
small → binds to mRNA
large → has peptidyl transferase activity and catalyzes formation of peptide bonds
what are the 3 distinctive tRNA binding areas on the ribosome
E, P, and A sites
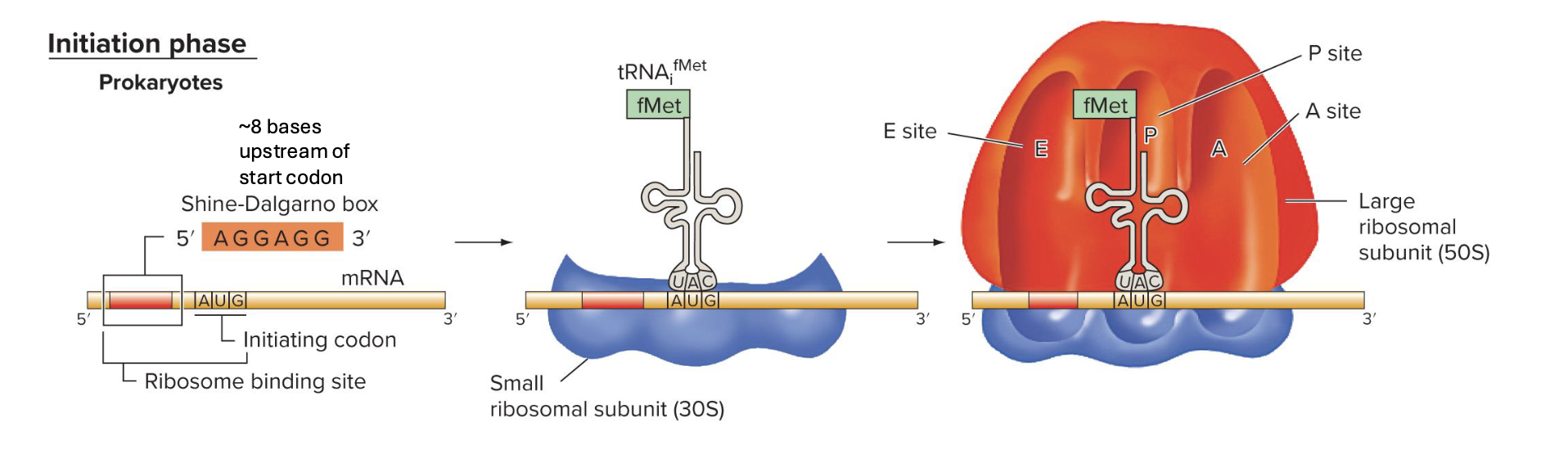
explain the initiation phase of translation in e. coli (prokaryotes)
Sine-Dalgarno sequence/box (on mRNA) is recognized by complementary sequences of 16 rRNA of the 30S subunit
fMet-tRNA is positioned in P site, complementary to AUG start codon on mRNA
large subunit binds
note: not all prokaryotes have same ribosome binding site recognition sequence as e. coli but many have sequences similar to it (have lots of purines (A and G))
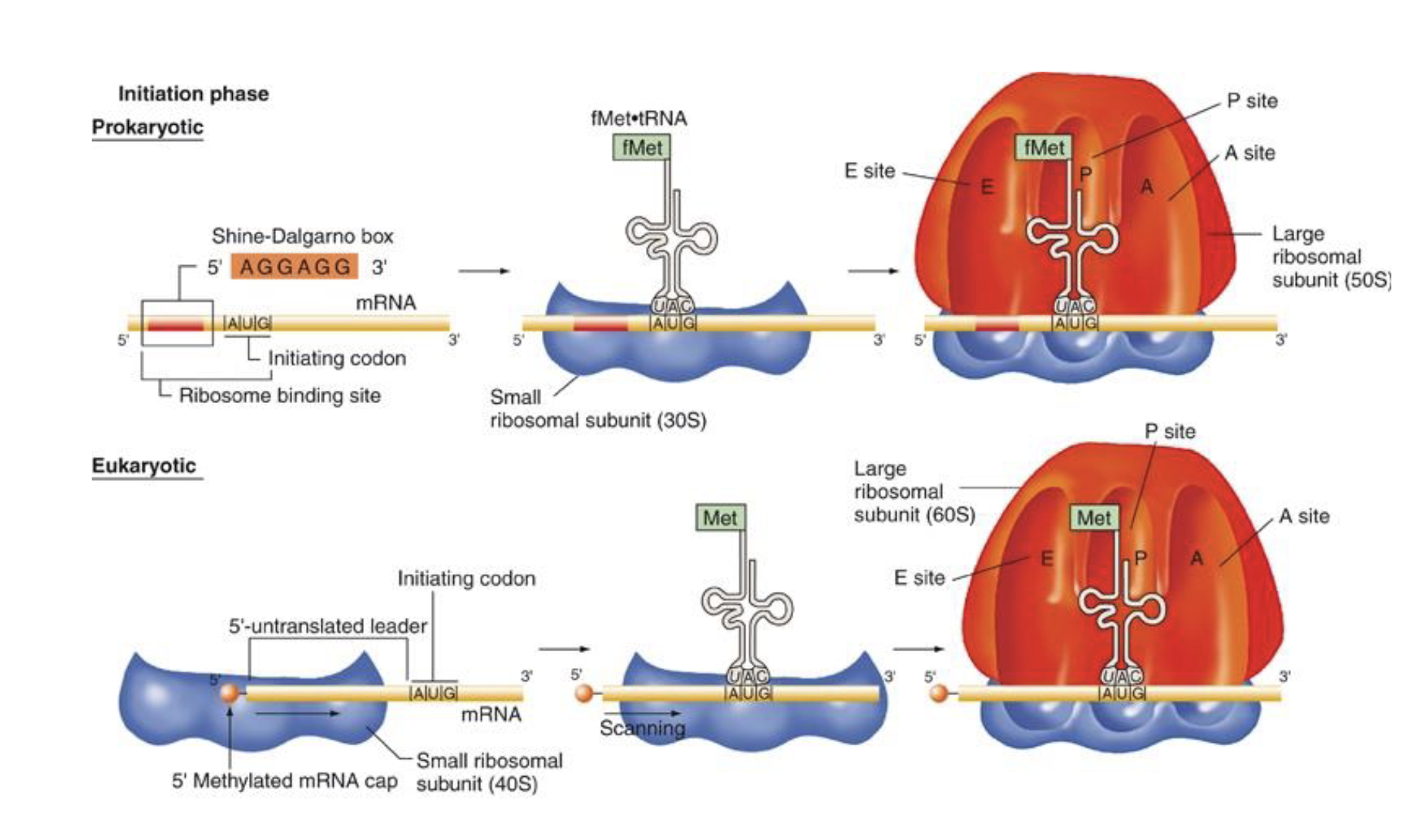
how does the initiation phase of translation in e. coli (prokaryotes) differ in eukaryotes
in Euks…
fMet is Met instead
ribosome binding site is the 5’ methyl cap in euks not the shine-dalgarno box
small ribosomal subunit is 40S not 30S
explain the elongation phase of translation in e. coli
elongation factors (EFs) escort next tRNA into the A site of the ribosome
peptidyl transferase (on large subunit) catalyzes formation of a peptide bond between carboxyl (C) terminus of formalmethionine (fMet) and the amino (N) terminus of the second AA
as ribosome moves, fMet moves from P to E site
new tRNA enters A site, fMet is released from E site
process repeats
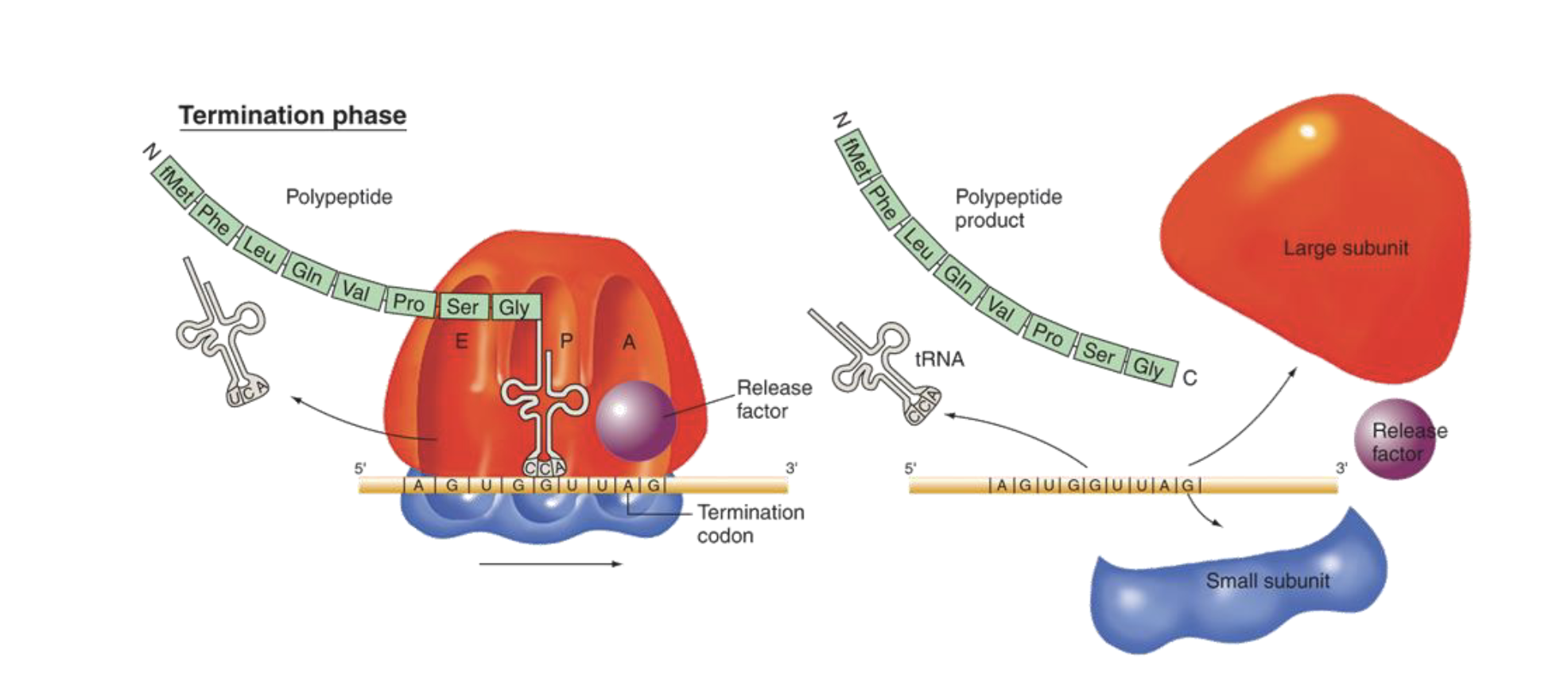
explain the termination phase of translation in e. coli
nonsense (stop) codon is encountered (for which there is no tRNA)
Release factor recognizes stop codon, moves into A site
polypeptide is released from C-terminal tRNA
mRNA, tRNA, and ribosomal subunits all dissociate from each other
what are polyribosomes
a complex of multiple ribosomes translating the same mRNA
allows simultaneous synthesis of many polypeptide copies from a single mRNA
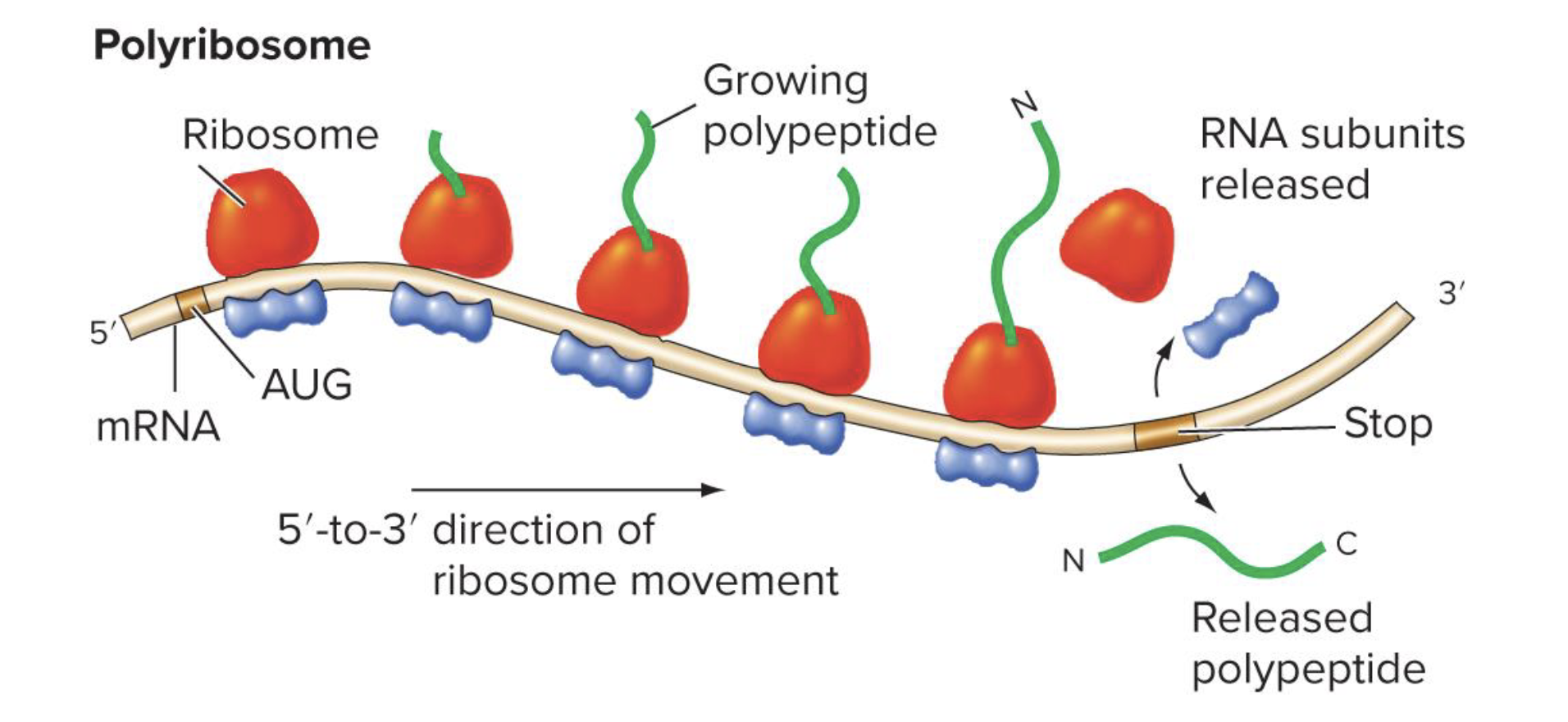
what is post-translational processing? give some exs
note: dont need to memorize exs
modifications that occur to protein after translation
eg enzymatic cleavage (remove bits and pieces of AA)
eg addition of chemical constituents (eg phosphorylation, glycosylaton, etc)
what are diffs in transcription/translation in pro and euks
proks:
no nucleus so transcription and translation happen in same cellular compartments (and translation is often couples to transcription)
genes are not divided into exons and introns
euks:
nuc is seperated from cytoplasm by a nuclear mem
transcription takes place in nucleus, translation in cytoplasm → so direct couping of transcription/translation is not possible
dna of gene consists of exons seperates by introns so the exons are defined by posttranscriptional splicing which deletes the introns
what are the diffs in prok and euk’s RNA polymerases
pro → has one RNA pol consisting of 5 subunits
euk → has 3 diff types of RNA pol, each w 10 or more subunits
diff rna pols transcribe diff types of genes
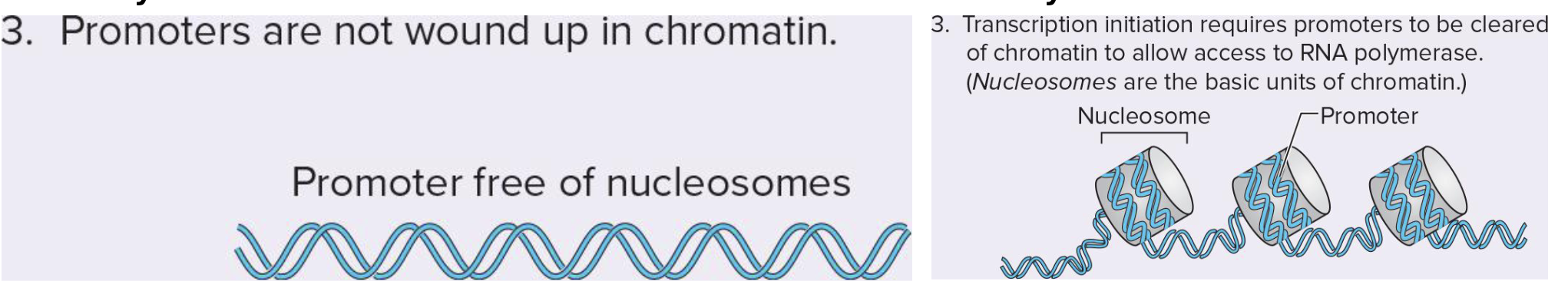
what are the diffs in prok and euk’s RNA promoters
proks → have accessible promoters
euks → promoters can be wound up as chromatin
enhancers clear histones from promoter and stabilize rna pol at the promoter
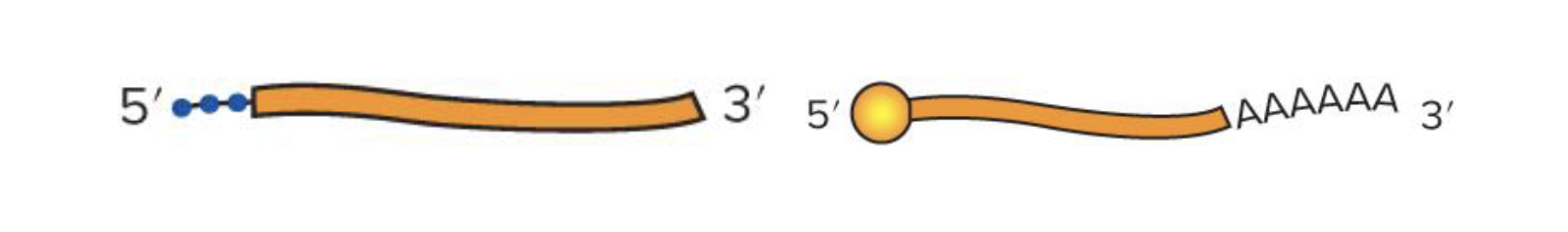
what are the diffs in the primary transcripts in proks and euks
proks → mRNA has triphosphate start at 5’ end and no tail at 3’ end
euks → primary transcript undergoes processing to produce mature mRNA
adding 5’ methyl cap and poly-a-tail
splicing out introns
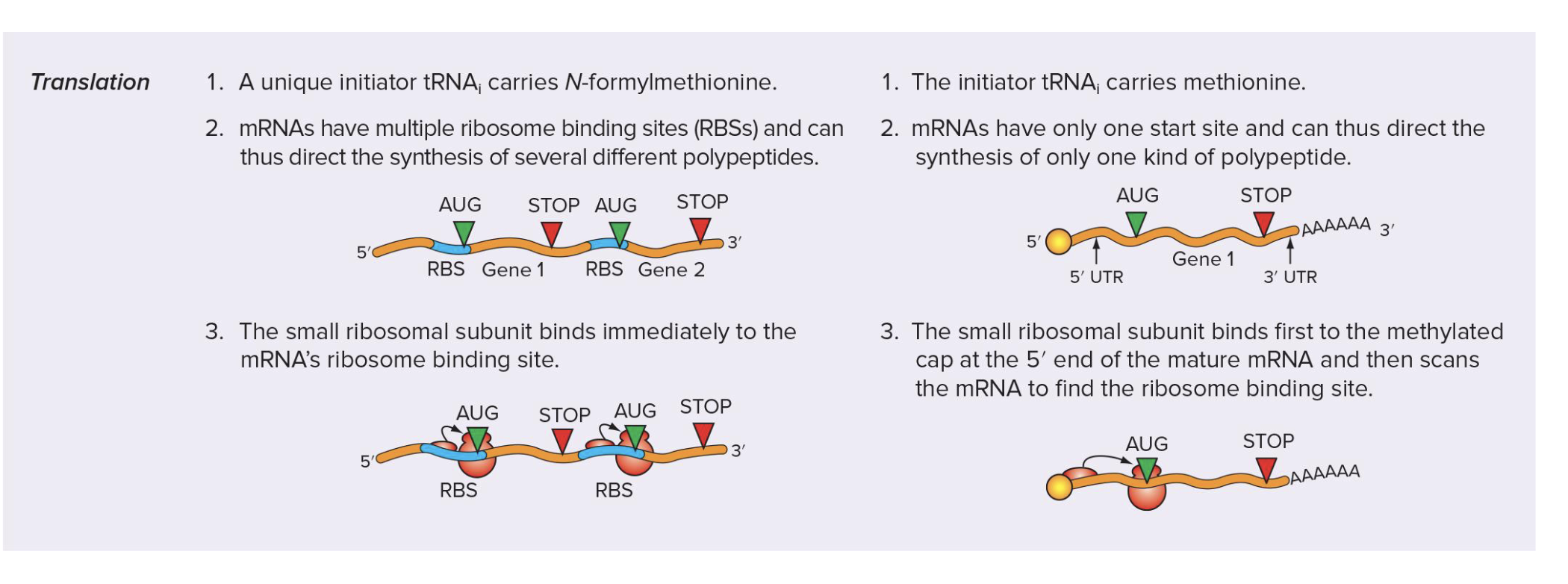
how does the initiation of translation differ between proks and euks
proks:
unique initiator tRNA carries N-formylmethionine (fMet)
mRNAs have multiple ribosome bining sites (RBSs) → direct synthesis of several diff polypeptides
small ribosomal subunit binds immediately to mRNA’s ribosome binding site
euks:
initiator tRNA carried methionine (Met)
mRNAs have only one start site → direct synthesis of only 1 kind of polypeptide
small ribosomal subunit binds first to methylated cap at 5’ end of mature mRNA and then scans mRNA to find ribosome binding site
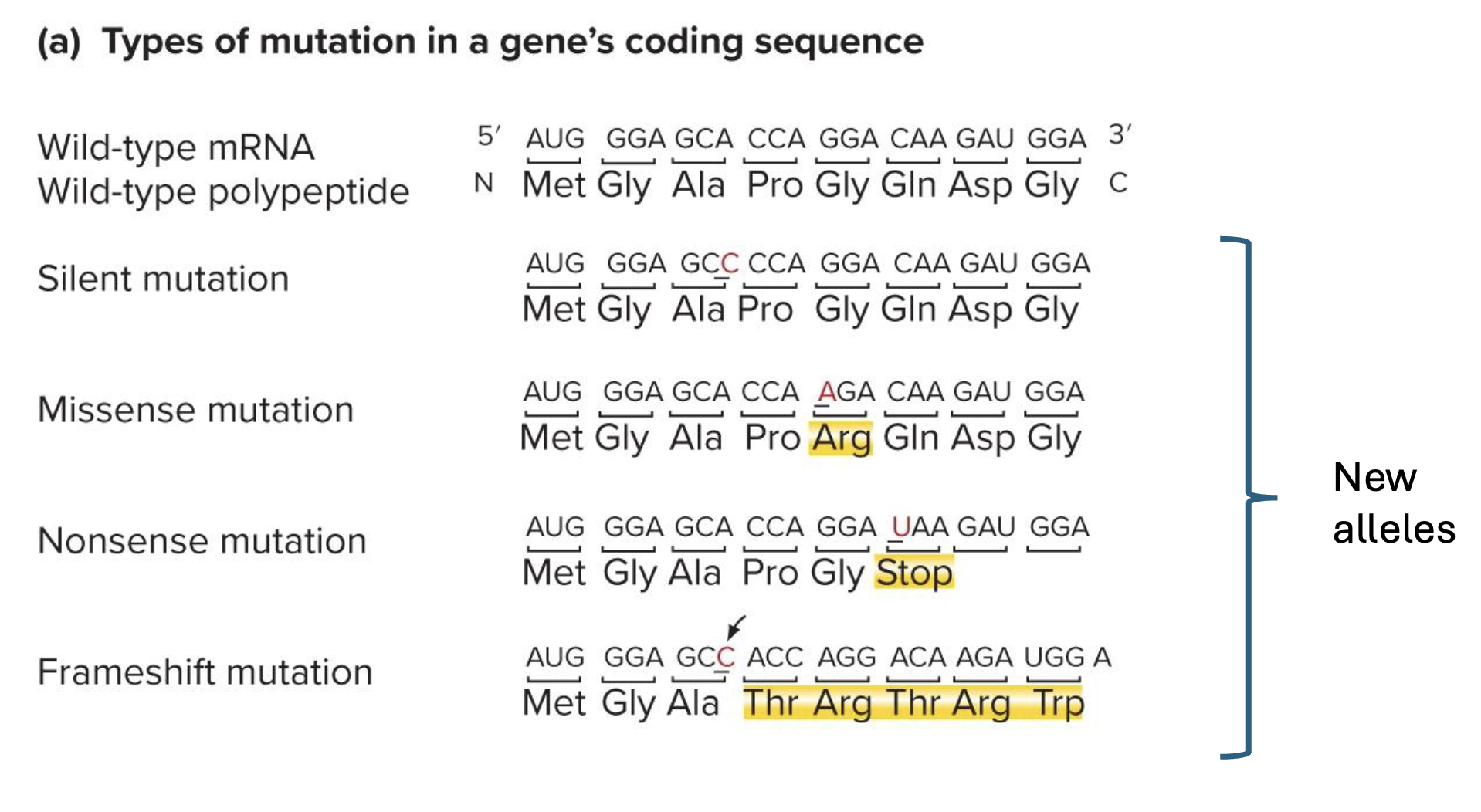
what are silent, missence, nonsense, and frameshift mutations? what do they all result in
all result in new alleles (even if silenc bc new sequence is diff)