AP Biology Review
1/87
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
88 Terms
Hydrolysis
Chemical reaction involving the addition of water to split a larger molecule into two smaller ones. This is a catabolic process.
Dehydration Synthesis
Chemical reaction involving the removal of a water molecule to combine two smaller molecules into a larger one. This is an anabolic process.
Adhesion
The attraction between water molecules and different substances, such as water adhering to plant cell walls. This property is crucial for processes like capillary action in plants.
Cohesion
The attraction between water molecules. This property contributes to surface tension and the formation of droplets.
Water Potential
(Ψ = Ψs + Ψp) This number always has to be less than zero. Water moves from areas of high water potential to low water potential, such as in a tree water moves upwards and evaporates because the water potential is lower higher up. This is measured in ‘bars.’
Solute Potential (Ψs)
This is determined by the concentration of solutes in a solution. The more solutes, the lower the solute potential (and thus the water potential), as water molecules are drawn to the solutes, making them less likely to move.
Ψs = -iCRT
i is the ionization constant (number of ions a solute dissociates into)
C is the molar concentration of the solute
R is the pressure constant
T is the temperature in Kelvin
Ionization Constant (i)
The number of ions a solute dissociates into when dissolved in a solution.
Ions like glucose and sucrose do not ionize in water, therefore i = 1. Whereas ions like NaCl dissolve into 2 making i = 2, and CaCl² i = 3.
Molar Concentration (C)
This is the moles/Liter of the solute in the water, or the molarity of the solution.
Pressure Constant (R)
This is always 0.0831 liter bars/mole K
Kelvin (T)
273 + C = T
Pressure Potential (Ψp)
Plants have cell walls, therefore their walls exert pressure on them, which is what this number is.
Osmosis
The movement of water across a semi-permeable membrane from an area of low solute concentration to an area of high solute concentration. Osmolarity is the total solute concentration in a solution.
Active Transport
The movement of ions or molecules across a cell membrane against their concentration gradient, requiring energy (typically from ATP).
Endocytosis- outside to inside
Phagocytosis- large particles
Pinocytosis- extracellular fluid with dissolved substances
Receptor-Mediated Endocytosis- receptor proteins on cell used to capture specific target molecules
Exocytosis- inside to outside
Proteins (signaling), Hormones, Waste
Passive Transport
The movement of substances across a cell membrane without the need for energy input, typically down their concentration gradient. NO ATP
Diffusion: molecules moving from high to low concentrations (small nonpolar)
Facilitated Diffusions: high to low through transport proteins, hydrophilic molecules and ions
Proteins
Protein function is dependent on its shape. Proteins have 4 levels of structure: primary, secondary, tertiary, and quaternary.
The sequence of amino acids in the polypeptide chain
The initial folding (alpha helices or beta sheets) of the polypeptide due to H-bonds
The interactions between the R groups of the amino acids
The overall 3D shape formed by multiple polypeptide chains
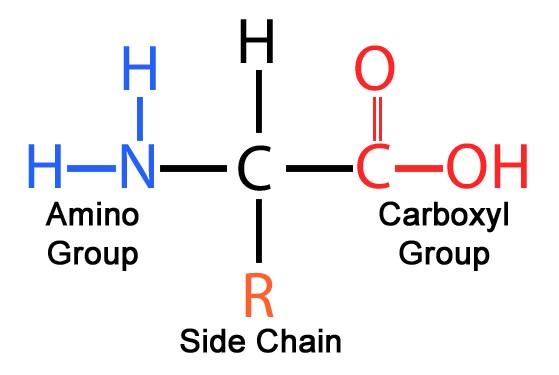
Amino Acids
Building blocks of proteins, consisting of a central carbon atom, an amino group, a carboxyl group, and a variable R group that determines their identity.
DNA
The carrier of genetic information in living organisms, composed of two strands that form a double helix structure, with sequences of nucleotides encoding the instructions for protein synthesis. It runs double parallel, with one strand in the 5’ to 3’ and the other in the 3’ to 5’ direction.
RNA
A nucleic acid that plays a crucial role in coding, decoding, regulation, and expression of genes. It is involved in protein synthesis and exists in several forms, including mRNA, tRNA, and rRNA. It does not have Thymine as a base and has Uracil instead, it is also made of a ribose base, not a deoxyribose base like DNA.
Ribosomes
They synthesize proteins according to mRNA, which originate in the genome of a cell, reflecting a common ancestry between prokaryotes and eukaryotes. Ribosomes have two subunits (not membrane enclosed) and are made of rRNA and proteins (tRNA).
Nucleus
Unique to eukaryotic cells, the nucleus is an organelle that houses the cell's genetic material, managing gene expression and coordinating cell activities such as growth and reproduction.
Endoplasmic Reticulum
Network of membrane tubules within cytoplasm of eukaryotes.
Rough ER
Ribosomes in membrane
Packages newly synthesized proteins for possible export from cell
Smooth ER
Lacks ribosomes
Functions are detox and lipid synthesis
Golgi Complex
Series of flattened membrane bound sacs in eukaryotes. Involved in proper folding and chemical modification of new proteins and packaging for protein trafficking (through transport vesicles).
Mitochondria
Has a double membrane and through cellular respiration and is known as the powerhouse of the cell as it produces ATP. Its outer membrane is smooth, inner is folded into cristae.
The Krebs Cycle (citric acid cycle) occurs in the matrix (inner)
Electron transport and ATP synthesis occur in inner mitochondrial membrane
Folding of the inner membrane increases surface area which allows for more ATP creation (through cristae folding)
Lysosomes
Contains hydrolytic enzymes to digest damaged cell parts or macromolecules.
Intracellular digestion
Apoptosis
Vacuoles
Membrane bound sacs in eukaryotes.
Used for storage (of water and macromolecules), and for the release of waste
Helps maintain turgor pressure
Chloroplasts
Found in eukaryotes like algae and plants, they capture energy from the sun and convert it into sugars. Has a double outer membrane.
Thylakoids
Highly folded membrane components stacked in grana
Containing chlorophyll pigments that comprise the photosystems and ETC proteins can be found between photosystems embedded in the thylakoid membrane
Light dependent reactions occur here
More folding = higher efficiency
Stroma
Fluid between inner chloroplast membrane outside thylakoids
Carbon Fixation reactions occur here
Surface Area to Volume Ratio
This is important because it limits the size of cells, as a larger SA:V ratio will yield in greater efficiency and transport/exchange of materials with the environment.
Adaptations for Better Exchange with Environment (SA:V)
Membrane Folding can be used to increase SA through
Villi and Microvilli on Small Intestine
Root Hairs on plants
Bigger animals have a harder time exchanging heat with their environment.
Leaves use stomata to increase exchange with environment.
Cholesterol
A type of steroid used to regulate the bilayer fluidity, wedged between phospholipids in the cell membrane of eukaryotes.
Phospholipids
Are amphipathic and have a hydrophilic, polar phosphate head, and a hydrophobic, nonpolar fatty acid tail to form a bilayer in aqueous environment.
Membrane Proteins
Peripheral Proteins
Loosely bound to surface
Hydrophilic with charge and polar side groups
Integral Proteins
Span the membrane
Hydrophobic with nonpolar side groups in the hydrophobic interior of bilayer and hydrophilic with charged and polar side groups on the exterior (like transmembrane proteins)
Functions
Transport
Cell-Cell Recognition
Enzymatic Activity
Signal Transduction
Intercellular joining
Attachment to extracellular matrix or cytoskeleton
Fluid Mosaic Model
The structure of the phospholipid bilayer is fluid and held together by hydrophobic interactions weaker than covalent bonds, allowing some lipids and proteins to flow along the surface of the membrane or across the bilayer.
Carbohydrates in the Fluid Mosaic Model
Both function as markers (cell markers)
Glycoproteins: one or more carbohydrates attached to membrane proteins
Glycolipids: lipid with one or more carbohydrates attached
Selective Permeability
Small Hydrophobic Molecules: can easily pass through membrane (ex: O2, CO2)
Large Polar, Hydrophilic Molecules: cannot move freely across the membrane and need the help of transport proteins:
Channel Proteins: hydrophilic tunnel allows specific target molecules to pass through
Carrier Proteins: spans the membrane and changes shape to move a target molecule from one side to the other
Cell Walls
Made of complex carbohydrates:
Plants: Cellulose (polysaccharide)
Fungi: Chitin (polysaccharide)
Prokaryotes: Peptidoglycan (polymer of sugar and amino acids)
Facilitated Diffusion
Movement of molecules from high to low concentrations through transport proteins, and example of active transport.
Aquaporins for water
Charged ions like Na+ and K+ (Carrier Proteins- Pumps) (can create membrane potential through the pumping of charged ions)
Large and small polar molecules
Cotransport, secondary active transport using energy from electrochemical gradient to transport two different ions across the membrane through a protein.
Symport- two different ions in same direction
Antiport- two different ions in opposite directions
Tonicity
Hypertonic Environment: more solute and less solvent than inside the cell (water will flow out of cell)
Isotonic: equal concentrations of solute and solvent to the cell
Hypotonic: less solute and more solvent than the cell (water will flow in)
Animal cells lyse (explode)
Plant cells are turgid
Paramecium use contractile vacuole to pump water out
Enzymes
Biological catalysts that speed up biochemical reactions (most enzymes are proteins and their shape is necessary for their function).
Active Site that interacts with substrates with a unique shape and charge (can be changed slightly to fit the substrate)
Enzyme names indicate chemical reaction or substrate (Ex: Sucrase digests sucrose)
Reusable, not changed by reactions
Activation Energy
Biochemical reactions need a starting energy called the activation energy
Some reactions result in a net release of energy, and some result in a net absorption of energy
Reactions resulting in a net release of energy need less activation energy than those resulting in a net absorption of energy
Enzymes lower the activation energy, thereby accelerating the rate of reactions
Controls
Negative control: not exposed to experimental treatment or any treatment known to have an effect
Positive control: exposed to a treatment with a known effect, not exposed to the experimental treatment
Denaturation
Changes in environmental temperature and pH can lead to denaturation (loss of enzyme shape), and therefore enzyme catalytic ability is lost or decreased
Higher temperatures result in denaturation, but lower temperatures generally just slow down the reaction rate without denaturation
Denaturation can occur at both increased or decreased pH because the change in hydrogen ion concentration can disrupt the H-bonds in enzyme structure
Most times this is irreversible
pH
pH measures the concentration of hydrogen ions in a solution. It is measured on a logarithmic scale so a pH of 6 has 10x more hydrogen ions in a solution than a pH of 7.
Low pH is acidic, high pH is alkaline or basic.
Inhibitors
Competitive Inhibitor
Can bind reversibly or irreversibly to active site of an enzyme
Competes with substrate for enzyme’s active site
Noncompetitive Inhibitor:
Bind to an enzyme’s allosteric site, not the active site, and thereby change the shape of the enzyme
Second Law of Thermodynamics
Every energy transfer increases the disorder of the universe
Living cells are not at equilibrium; there is a constant flow of materials in and out of them
Cells manage energy resources through energy coupling (energy releasing processes drive energy storing processes)
Photosynthesis: Light Dependent Reactions
Light-dependent Reactions: capture light energy through pigments
Pigments transform light energy into chemical energy
Chlorophyll: capture sun energy into high-energy electrons
Energy from these electrons is used to establish a proton gradient and reduce NADP+ to NADPH
Chemical energy is temporarily stored in the chemical bonds of carrier molecules called NADPH
Help facilitate ATP synthesis
ATP and NADPH (products of light-dependent) store chemical energy to power the Calvin Cycle (will be reactants)
Oxygen is produced as a result of water hydrolysis
Thylakoid Membrane and the ETC:
Photosystem (PS) II (is actually first) and uses hydrolysis of water and releasing of hydrogen molecules released into the thylakoid space to create a proton gradient
ATP synthase at the end of the ETC uses the proton gradient to turn ADP into ATP
Photosynthesis: Light-Independent Reactions (The Calvin Cycle)
The Calvin Cycle: uses ATP, NADPH, and CO2 to make glucose.
Carbon Fixation- Stroma: CO2, enzyme RuBisCO, and RuBP (five atoms of carbon and a phosphate group on each end)
RuBisCO catalyzes a reaction between CO2 and RuBP, which forms a six-carbon compound that is immediately converted into two three-carbon compounds.
Reduction: ATP and NADPH use their stored energy to convert the three-carbon compound, 3-PGA, into another three-carbon compound called G3P. (involves the gain of electrons) The molecules of ADP and NAD+, resulting from the reduction reaction, return to the light-dependent reactions to be re-energized.
Regeneration:
One of the G3P molecules leaves the Calvin cycle to form glucose (C6H12O6), because it has 6C, it takes six turns of the Calvin cycle to make one
The remaining G3P molecules regenerate RuBP, which enables the system to prepare for the carbon-fixation step. ATP is also used in the regeneration of RuBP.
Fermentation- Anaerobic Respiration
Glucose is broken down through glycolysis and uses 2 ATP to create pyruvate. If oxygen is not present at this point, fermentation occurs to create either lactic acid or ethanol.
Cellular Respiration
Glucose is broken down through glycolysis (in cytoplasm) and uses 2 ATP to create pyruvate. If oxygen is present at this point, cellular respiration occurs.
Glycolysis— in the cytoplasm
Results in pyruvate, NADH, and ATP
2 pyruvate per one molecule of glucose is then transported actively through the mitochondrial membranes into the matrix
Pyruvate Oxidation— in mitochondria
Products are NADH+ which becomes NADH and H+, CO2, and Acethyl CoA
Acethyl CoA then enters the Krebs Cycle
Krebs Cycle (Citric Acid Cycle)— in mitochondria
High energy electrons are transferred to NADH and FADH2
ADP is phosphorylated forming ATP
ETC— in inner mitochondrial membrane and internal membrane of chloroplasts (in prokaryotes— ETC is in plasma membrane)
Oxidative Phosphorylation
Electrons delivered by electron carriers (NADH and FADH2 to the ETC)
Use of Active Transport
ATP synthase uses proton gradient to synthesize ATP
Decoupling oxidative phosphorylation (proton gradient not used by ATP synthase) is energy released as heat used by endothermic organisms to regulate body temperature
Creation of up to 36 ATP
Short Distance Cell Communication
Cell sounds out local regulators (signals), and the target cell is within a short distance of the signal (local signaling). This is often used to communicate with cells of the same type.
Long Distance Cell Communication
The target cell is not in the same area as the cell emitting the signal, and so the signal travels a long distance to reach the target cell, which is usually a cell of a different type.
Immune Cell Communcation
They communicate through cell-to-cell contact. The B-cell is triggered by the binding of specific antigens to the B-cell receptor, which sets off further signals.
Neurotransmitter Communication
Exocytotic vesicles release signals and those signals travel short distances across small gaps between cells, then attach to the target cell.
Signal Transduction Pathway (RTR)
Reception of the signal from outside the cell
Transduction converts the signal to form that can obtain a cellular response (signal transduction pathway)
Phosphorylation cascade
Response
Regulation of protein synthesis by turning genes in the nucleus on/off
Regulation of protein activity in cytoplasm
Chemicals can either inhibit or activate pathways (such as blocking neurotransmitters, or activation of a receptor protein)
Transduction In Depth
Ligand (chemical messenger- hormone) binds to the receptor on the outside of the cell membrane (ex: G protein-coupled receptors in eukaryotes)
GDP —> GTP (G protein coupled receptor is activated by ligand
GTP then activates Adenylate cyclase and ATP is released
Which is then converted to cAMP (second messenger) which activates protein kinases which amplify the signal
Sidenote- steroid transduction is different because the steroid simply passes through the membrane then enters the nucleus immediately to activate the production of a protein
Quorum Sensing
Signal mechanism used by bacteria where they release signals to their neighbors. This type allows bacteria to sense changes in population density and they can then act as a group to respond to environmental change.
Positive Feedback
Amplify responses and processes, one variable initiates a response and this disrupts homeostasis and moves further and further from it. (creates more change) Amplifies the change.
Example: Ethylene release and ripening of apples, birth
Negative Feedback
If a system is disrupted, negative feedback mechanisms works to reset it back to its normal state, maintaining homeostasis.
The Cell Cycle
Interphase: growth and preparation
G1: cell growth
S: DNA replicated
G2: cytoplasmic components doubled
M-Phase: division of nucleus and cytoplasm, results in TWO GENETICALLY IDENTICAL DAUGHTER CELLS
Mitosis: growth, repair, asexual reproduction
Prophase:
Nuclear envelope begins to disappear
DNA coils into chromosomes
Fibers move double chromosomes toward center of cell
Metaphase:
Fibers align double chromosomes across cell
Anaphase
Fibers separate double chromosomes into single ones (chromatids) at the centromere
Migration of chromatids to opposite sides of cell
Telophase:
Nuclear envelope reappears
Each nucleus contains a complete genome and chromosomes begin to uncoil
Cytokinesis: division of cytoplasm
G0 Phase: no more cell division
Cell Cycle Checkpoints
G1 Checkpoint: at end of G1 Phase
Cell size check
Nutrient check
Growth Factor check
DNA damage check
G2 Checkpoint: at end of G2
DNA replication check
DNA damage check
M-spindle Checkpoint: at end of metaphase
Fiber attachment to chromosome check (for proper division)
Cyclins and Kinases
Cyclins: group of related proteins associated with specific cell cycle phases
different cyclins at different stages
concentrations fluctuate on cell activity
used to promote cell cycle progression—> degraded to inhibit cycle progression
used to activate cyclin-dependent kinases (CDKs) (cyclins are specific to their CDK)
CDKs: group of enzymes in cell cycle regulation
requires cyclin binding for activation
phosphorylate substrates and promotes certain cell cycle activities
Meiosis
Haploid n: gametes for sexual reproduction (one set of chromosomes)
Diploid 2n: body cells (2 full sets of chromosomes)
Produces 4 haploid daughter cells
Meiosis I:
Prophase I:
Nuclear envelope disappears, fibers form, DNA coils into sister chromatids
Double chromosomes pair up and exchange genetic information (crossing over- forms recombinant chromatids)
Metaphase I
Sister chromatids remain in pairs, fibers align pairs across center of cell
Random assortment occurs = affects which chromosomes end up in each gamete = increases variation
Anaphase I
Fibers separate chromosome pairs
Each sister chromatid from the pair migrates to opposite sides of the cell
Telophase I
Nuclear envelope reappears and establishes separate nuclei with only ONE double chromosome from each pair = half of total info that the parent contained but still double chromosomes (sister chromatids)
Chromosomes uncoil
Cytokinesis occurs and forms two daughter cells which are haploid and genetically different
Meiosis II:
Prophase II:
Nuclear envelope disappears, fibers form
Metaphase II:
Fibers align sister chromatids across center of cell
Anaphase II:
Fibers separate sister chromatids
Chromatids move to different sides of the cell
Telophase II:
Nuclear envelope reappears and establishes separate nuclei with only a single chromosome
Cytokinesis occurs and forms four genetically different daughter cells
Phenotype Plasiticty
Environmental factors can change the expression of genes, and phenotypic plasticity is the ability of one genotype to produce multiple phenotypes.
Ex: Hydrangea Plant color is determined by the pH of the soil
Ex: Stomata density dependent on CO2 levels in atmosphere
Independent Assortment
Genes will be sorted into gametes independently and they are not linked. Genes are linked if they have a map unit lower than 50.
Nondisjunction
The failure of chromosomes to fully separate during the formation of gametes = too few or too many chromosomes in the sex cells. Ex: Down Syndrome, or Trisomy 21.
Ex: Triple X syndrome
Pyrimidines
Uracil, Cytosine, Thymine = one ring structure
Purines
Adenine, Guanine = double ring structure
DNA Replication
Semiconservative replication: resulting DNA contains one original (template) strand and one newly synthesized complimentary strand.
Directionality of DNA strands: antiparallel
Phosphate terminus is 5’
Hydroxyl (OH) terminus is 3’
New nucleotides can only be added in the 5’—3’ direction so one strand is synthesized continuously (leading strand) and the other (lagging strand) is synthesized in fragments
Helicase: unwinds DNA strands
Topoisomerase: relaxes supercoil at replicatio fork- where two strands are separated
DNA Polymerase: synthesizes new strands
Needs RNA primers to start synthesis
Attaches to the 3’ end of the template strand and builds in 5’ to 3’ direction
Ligase: joins DNA fragments on lagging strand
Transcription
DNA strands are separated and the template strand (noncoding strand) is transcribed by RNA polymerase in the 5’ to 3’ direction by reading in the 3’ to 5’ direction and creates mRNA of one particular gene.
RNA Processing/Modification
Addition of Poly-A tail (3’) and GTP head (5’)
Introns are excised
Exons are retained
Alternative Splicing:
Produce different mRNA transcripts from one primary transcript through keeping of different exons
Retrovirus Translation
Introduction of viral RNA (not DNA) into host cells through the use of the enzyme reverse transcriptase that copies viral RNA into viral DNA, once it converts viral RNA to viral DNA, it is integrated into the host genome and will then be transcribed and translated, this results in the assembly of new viral progeny.
Regulatory Sequences
Stretches of DNA used to promote or inhibit protein synthesis by coding for regulatory proteins = controls transcription.
Epigenetics
Epigenetic changes are reversible modification of DNA or histones (proteins used to wrap DNA around) with slight chemical changes that cause histones to tightly or loosely pack DNA = gene expression regulation with access to DNA.
Transcription Factors
Proteins that promote or inhibit transcription of a gene = determines how a cell differentiates during development.
Operons
Closely linked genes producing a single mRNA molecule during transcription under the control of the same regulatory sequence (includes genes to be transcribed, regulatory sequence, and operator).
Operator either inhibits or promotes
Ex: Lac Operon is inducible (usually off, with regulatory protein bound to operator and RNA polymerase cannot bind to DNA)
Inducers are molecules that bind to regulatory protein and cause it to change shape and unbind or be unable to bind to the operator (Ex: allolactose in Lac Operon)
Small RNA fragments (sRNA)
can break down mRNA in cytoplasm as a form of gene expression
can block transcription as well
Mutations in Chromosome Number
Triploidy: 3 copies of a particular chromosome = sterility in some animals
Polyploidy: multiple sets of homologous chromosomes = can result in increased vigor in plants
Trisomy 21: extra chromosome at number 21
Turner Syndrome: missing X chromosome
Horizontal Genetic Transfer
Exchange of genetic info between different genomes or unrelated organisms. Is mainly seen in prokaryotes and contributes to genetic variation in prokaryotes through
Transformation: DNA comes from external environmental source (naked)
Transduction: transmission of foreign DNA into a cell when viral genome integrates with host genome
Conjugation: cell to cell transfer of DNA
Transposition: movement of DNA segments within or between DNA molecules
Gel Electrophoresis
Separates molecules according to size and charge. DNA is negatively charged and DNA samples with fragments are loading into wells with gel.
Current is then applied to gel creating positive and negative ends, as DNA moves to the positive end of gel it is separated by size and smaller particles move faster and further throughout the gel.
The resulting bands indicate molecules that migrated to that location. These band patterns can be used to identify people.
Polymerase Chain Reaction (PCR)
PCR allows the creation of large samples of DNA through DNA denaturing, then primers are added and DNA is replicated until an ample amount of DNA is obtained.
Convergent Evolution
Similar environmental conditions select for similar traits in different populations/species over time = analogous structures
Genetic drift
Random change in the frequency of a particular allele within a population. Like natural catastrophes, etc.
Bottleneck Events: sudden population decrease
Founder Effect: separation/isolation of population
Migration, geologic events
Migration/gene flow = movement of individuals between populations leading to an exchange of alleles between populations
Hardy Weinburg Equilibrium
Allele frequencies must stay constant (no genetic drift or gene flow)
p²+2pq+q²=1
p+q=1
Prezygotic Barriers
Habitat Isolation: species occupy different habitats and rarely come into contact
Temporal Isolation: species breed during different times of day, seasons, or years
Behavioral Isolation: species have different courtship rituals or mate preferences
Mechanical Isolation: reproductive structural differences prevent successful mating and reproduction
Gamete Isolation: sperm of one species may not be able to fertilize the eggs of another species
Postzygotic Barriers
Hybrid Inviability: Mating results in a zygote but incompatibility may stop its development
Hybrid Sterility: A hybrid offspring is produced that is vigorous but may be sterile
Hybrid Breakdown: First-generation hybrids are viable and fertile but resulting generations are feeble or sterile
Allopatric Speciation
Evolution of new species due to individuals from the same population being geographically isolated over a long period of time with no gene flow and different selective pressures.
Sympatric Speciation
Evolution of a new species due to individuals being reproductively isolated from a surviving ancestral population with no geographic barrier but instead genetic mutations, habitat differentiation or sexual selection.
Divergent Evolution
Adaptation to new habitats result in phenotypic diversification. Adaptive radiation refers to the evolution of new species that allows empty ecological roles or niches to be filled (such as after an extinction event).