Chapter 20: DNA Technology and Biotechnology Applications
1/274
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
275 Terms
DNA sequencing
A process used to determine the complete nucleotide sequence of a gene or DNA segment.
Plasmid
A small, circular, double-stranded DNA molecule that carries genes separate from those of a bacterial chromosome and can replicate separately as well.
Recombinant DNA
A molecule that contains DNA from two different sources.
Restriction enzymes
A type of enzyme used in gene cloning and genetic engineering that cuts DNA molecules at a limited number of specific locations.
Gel electrophoresis
This is a technique that incorporates a gel made of a polymer that acts as a molecular sieve to separate nucleic acids or proteins differing in size, electrical charge, or other physical properties. The gel has microscopic holes of different sizes that allow nucleic acids or proteins with shorter fragments to travel faster.

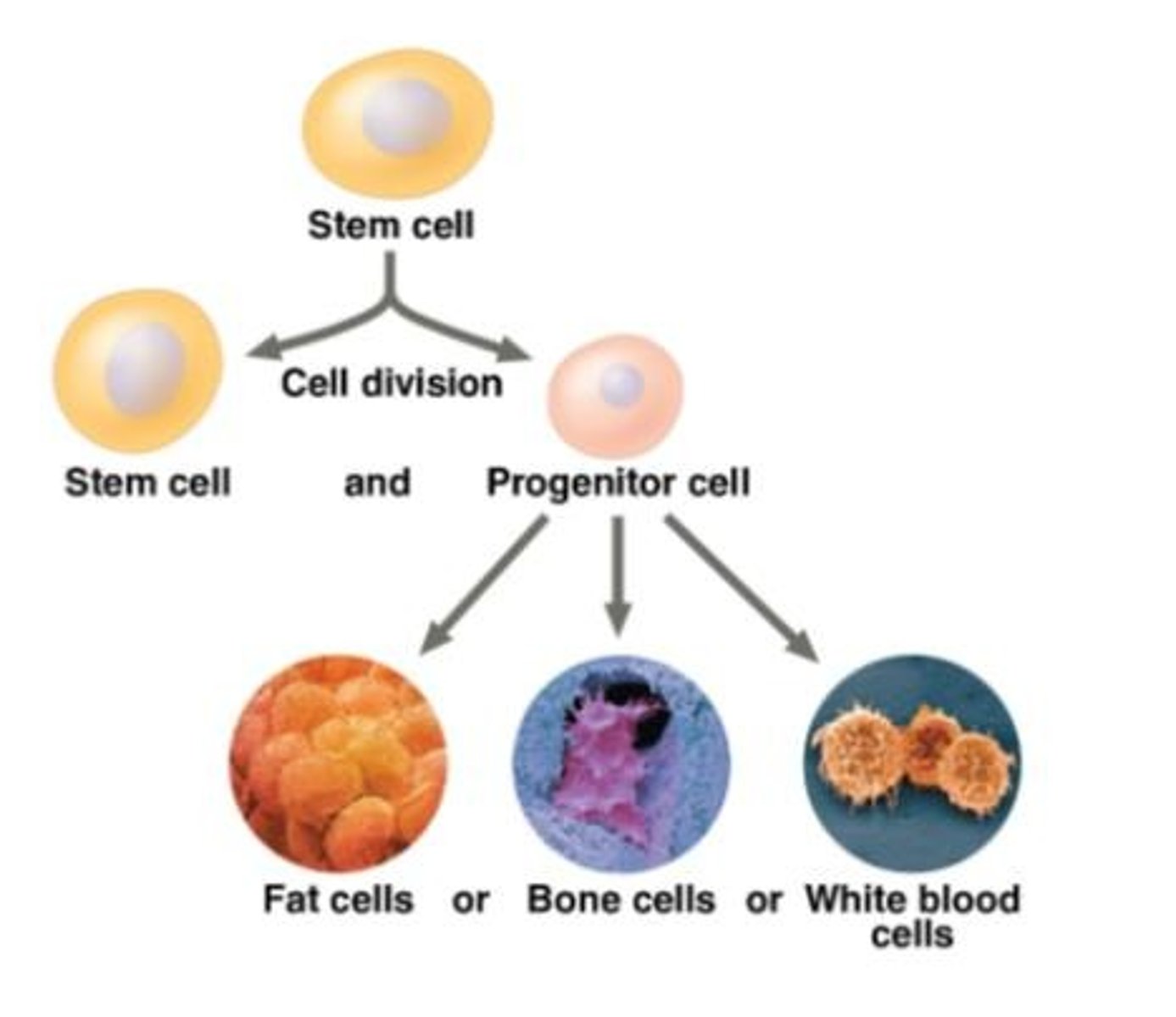
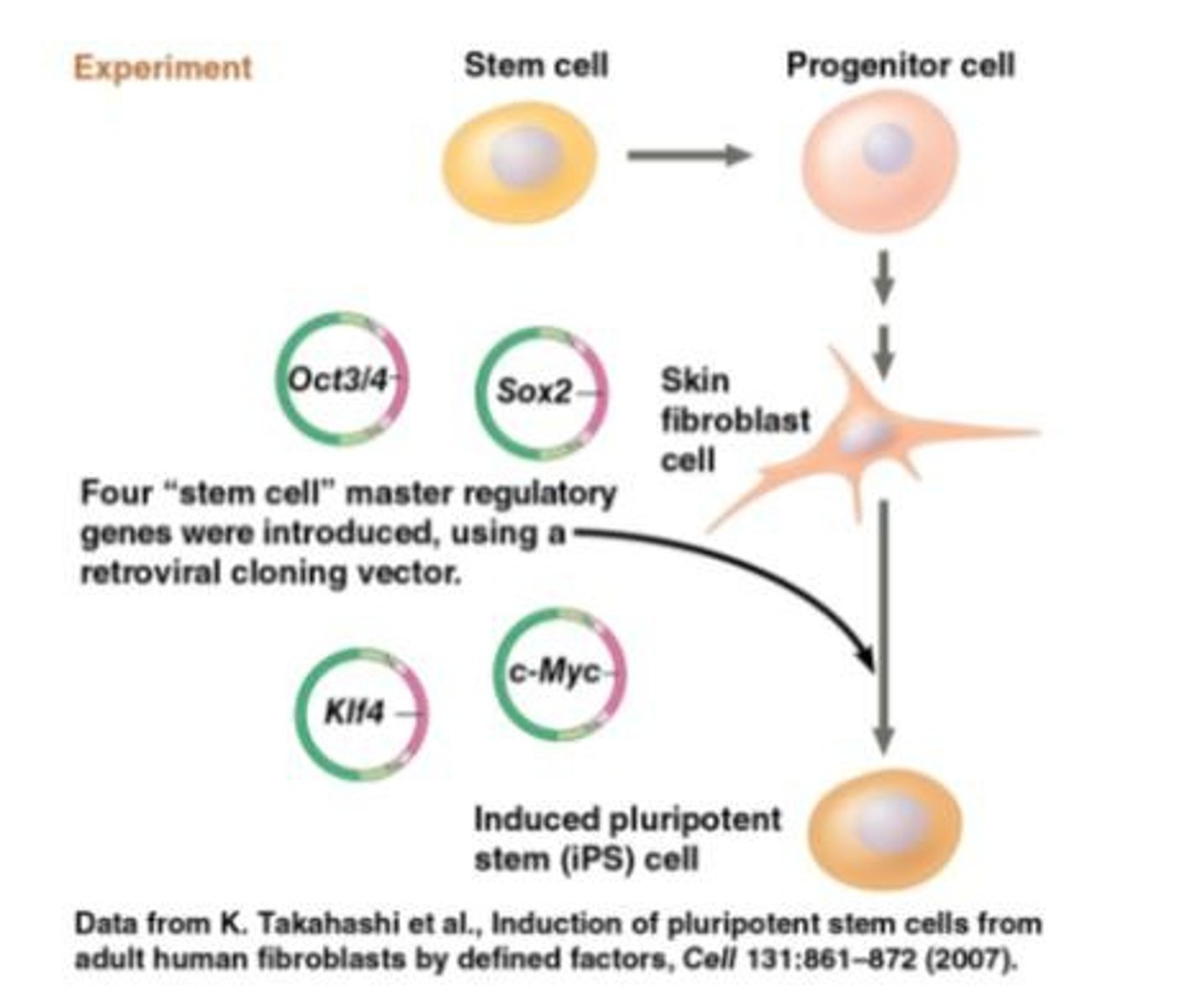
Stem cell
A relatively unspecialized cell that can reproduce itself indefinitely, or under certain conditions can differentiate into one or more types of specialized cells.
Gene therapy
The introduction of genes into an afflicted individual for therapeutic purposes. This holds great potential for treating disorders traceable to a single defective gene.
Gene cloning
The preparation of multiple identical copies of well-defined segments of DNA.
Polymerase chain reaction (PCR)
The main three steps of the PCR cycle include denaturing, annealing, and extension.
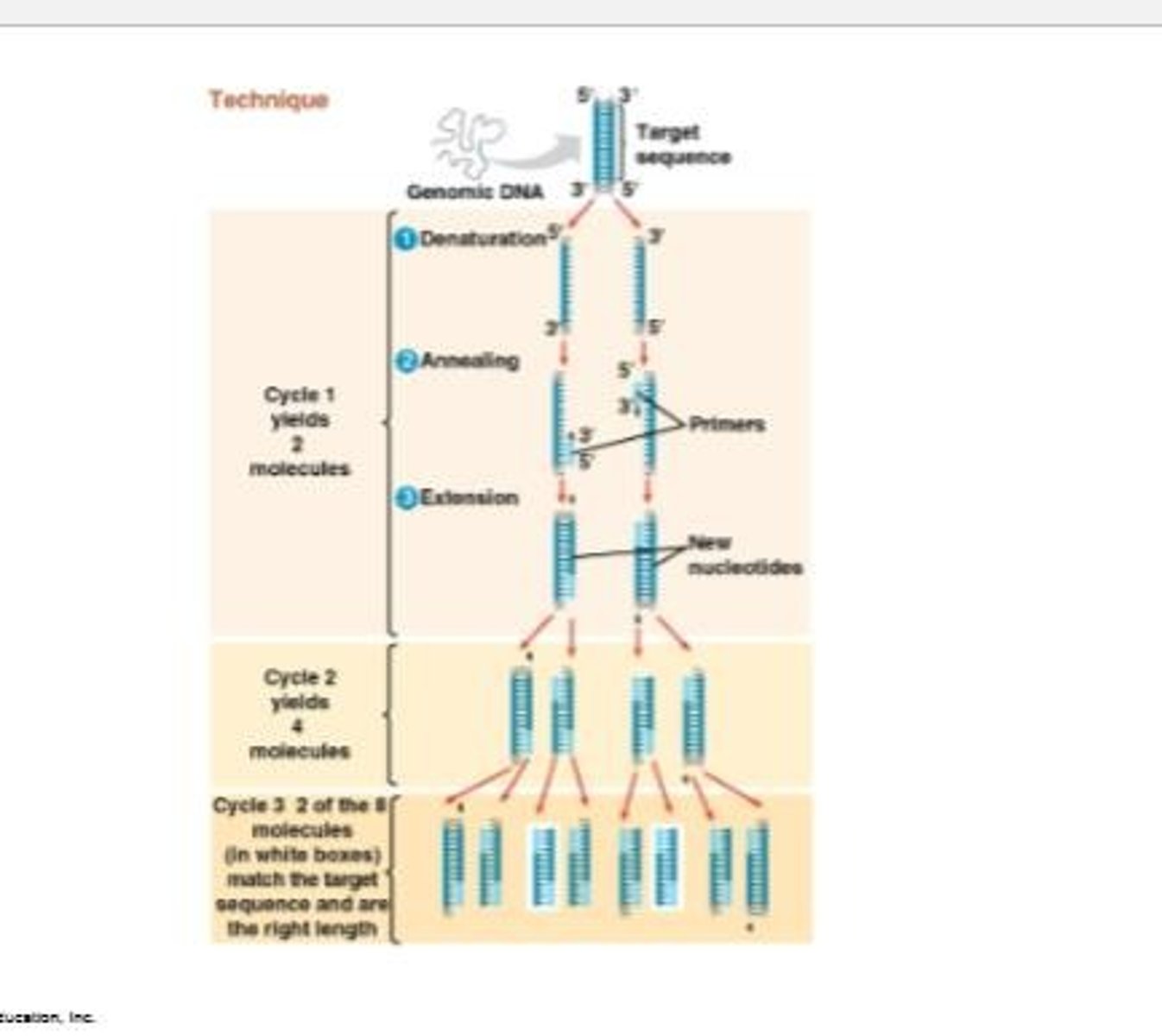
Denaturing
The first step in PCR where double-stranded DNA is heated until the strands are separated.
Annealing
The second step in PCR where separated strands are cooled to allow the annealing of short, single-stranded DNA primers that are complementary to sequences on opposite strands.
Extension
The final step in PCR where a heat-stable DNA polymerase extends the primers in the 5' → 3' direction.
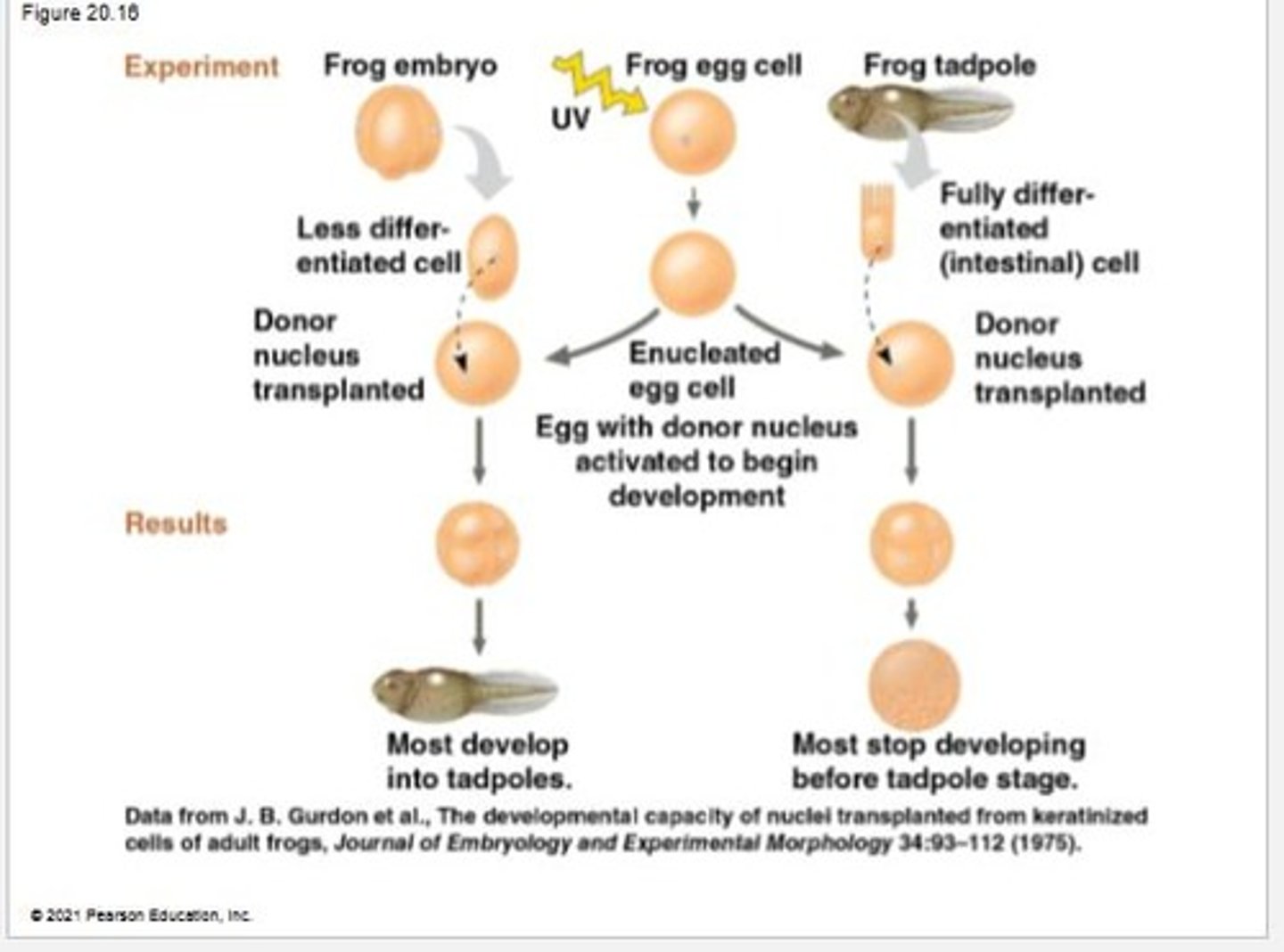
Nuclear transplantation
The process of replacing the nucleus of an egg with the nucleus of a differentiated donor cell.
Embryonic development
The process initiated after replacing the nucleus of an egg with the nucleus of a cell of the animal they wish to clone.
Advantage of stem cells in therapy
Stem cells can both reproduce indefinitely and differentiate into specialized cells needed for treatment, allowing replacement of damaged or diseased tissue.
Nanopore sequencing
A modern method of DNA sequencing that involves a membrane with a nanopore, where a single DNA strand is pulled through the pore one base at a time, causing unique disruptions in an ionic current that are measured and decoded.
Real-time DNA sequencing
A capability of nanopore sequencing that allows for immediate sequencing of DNA as it passes through the nanopore.
Portability of nanopore sequencer
The nanopore sequencer is almost the size of a small candy bar and connects to a computer via a USB port.
Cost-effectiveness of nanopore sequencing
Nanopore sequencing can be done in 6 hours on a device that costs $900.
DNA technology
The main technologies for sequencing and manipulating DNA, including techniques like DNA sequencing and gene cloning.
Nucleic acid hybridization
The base pairing of one strand of nucleic acid to the complementary sequence on another strand, based on the complementarity of the two DNA strands.
Genetic engineering
The direct manipulation of genes for practical purposes.
Dideoxy sequencing
An automated DNA sequencing technique developed by Frederick Sanger that uses a template strand to synthesize a nested set of complementary fragments.
Next-generation sequencing
Rapid and inexpensive sequencing techniques developed in the first decade of this century that can sequence 70-90 million nucleotides per hour.
Sanger method of DNA sequencing
A reaction involving template DNA, primer, DNA polymerase, nucleotides, and fluorescently-labeled dideoxynucleotides to determine the DNA sequence.
Denaturation in Sanger sequencing
The process where DNA is heated to separate the strands before cooling to allow the primer to bind.
Role of DNA polymerase
An enzyme that lengthens the DNA strand according to the template until a dideoxynucleotide is randomly placed, terminating the chain.
Dideoxynucleotide
A nucleotide missing the hydroxyl group that prevents further nucleotide addition, thus terminating the DNA chain.
Color association in Sanger sequencing
The color of the dideoxynucleotide is associated with the last base added to the DNA fragment.
Fragment separation in Sanger sequencing
The process of separating DNA fragments by size to read the dyes and determine the sequence.
PCR amplification
A technique used to amplify specific DNA sequences, making millions of copies of a particular segment.
Expressing genes
The process of producing the protein for which a gene codes.
Analyzing gene expression
The study of how genes are expressed and regulated in different conditions.
Gene editing
The process of making precise changes to the DNA of an organism.
Applications of biotechnology
Includes agriculture, medicine, environmental cleanup, ancestry, and forensic analysis.
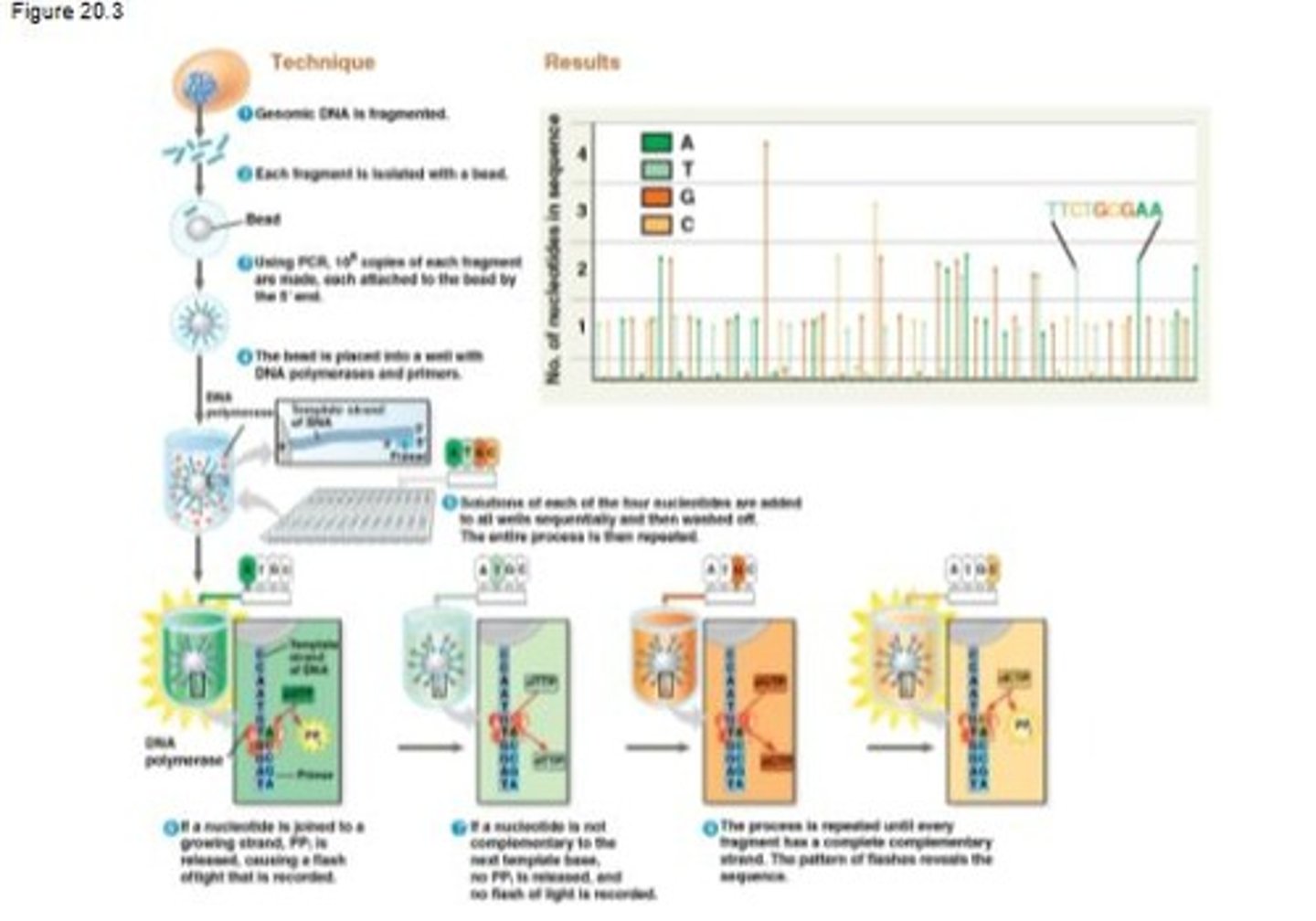
Sequencing by synthesis
A technique where a single strand of each DNA fragment is immobilized and the complementary strand is synthesized one nucleotide at a time.
High-throughput technology
A technology that allows thousands or hundreds of thousands of DNA fragments, about 300 nucleotides long, to be sequenced in parallel.
Genome DNA fragmentation
The process of breaking genomic DNA into smaller fragments, typically around 300 base pairs.
PCR (Polymerase Chain Reaction)
A technique used to make 10^6 copies of each DNA fragment, attaching them to a bead by the 5' prime end.
Bead isolation
The process of isolating each DNA fragment with a bead in a droplet of aqueous solution.
DNA polymerase
An enzyme used in the sequencing process that synthesizes new DNA strands complementary to the template strand.
Nucleotide addition
The process of adding solutions of each of the four nucleotides to the wells containing DNA fragments.
dNTPs (deoxynucleotide triphosphates)
The four nucleotides required for DNA synthesis: dATP, dTTP, dGTP, and dCTP.
Flash of light in sequencing
A signal recorded when a nucleotide is joined to a growing strand, indicating a complementary match.
Non-complementary nucleotide
A nucleotide that does not match the next base on the template strand, resulting in no flash of light being recorded.
Complete complementary strand
A strand that is fully synthesized to match the original DNA fragment, revealed by the pattern of flashes.
Third generation sequencing
A faster and less expensive sequencing method that sequences a single long DNA molecule as it moves through a nanopore.
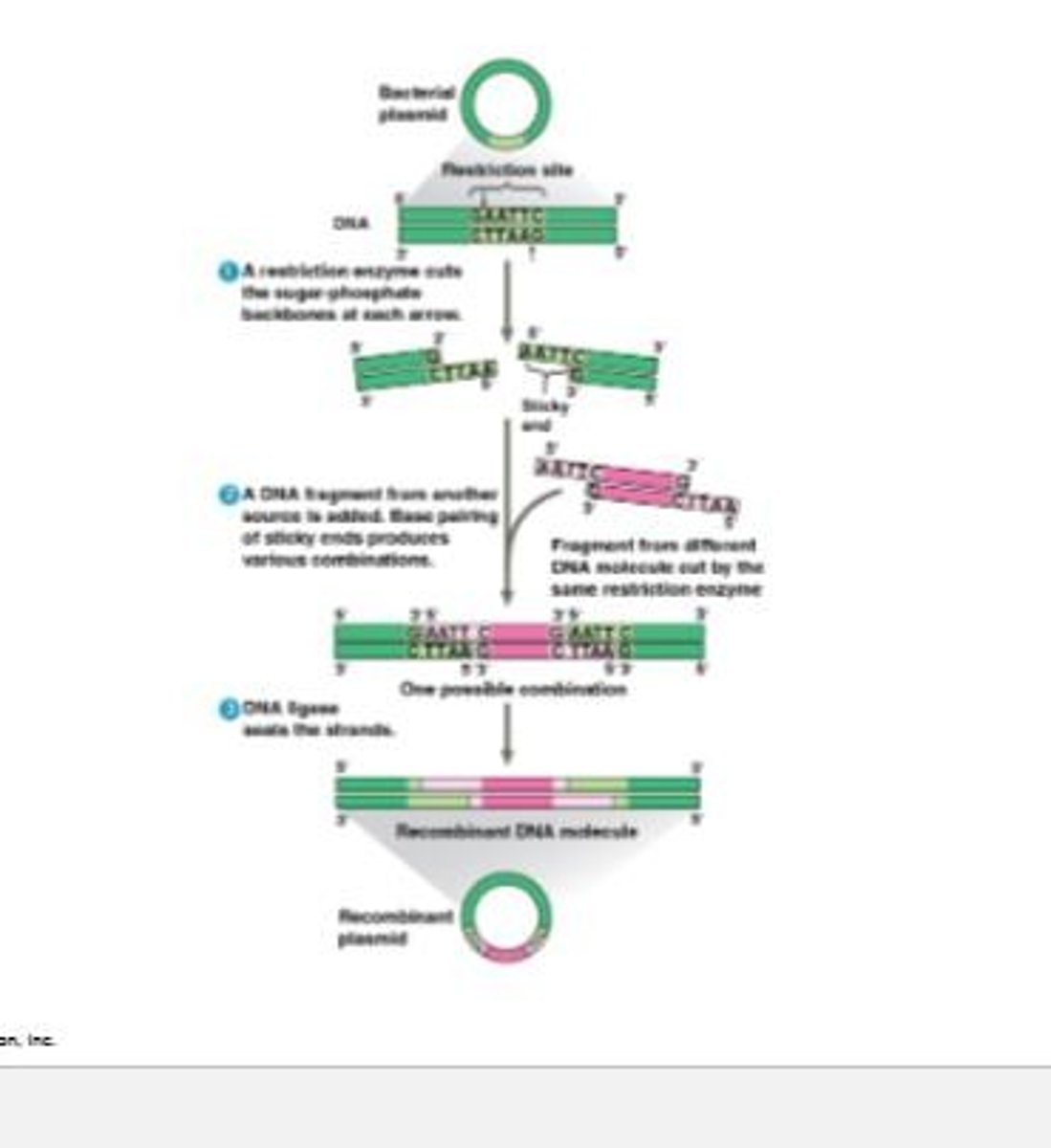
DNA cloning
A process used to prepare well-defined DNA segments in multiple identical copies for working directly with specific genes.
Plasmids
Small DNA molecules that replicate on their own and are used as vectors in DNA cloning.
Recombinant DNA molecule
A DNA molecule that contains DNA from two different sources, created by inserting a gene of interest into a plasmid.
Gene insertion into plasmid
The process of extracting a plasmid from a bacterial cell and inserting a gene of interest to create recombinant DNA.
Vector
A vehicle, such as a plasmid, used to transfer genetic material into a host cell.
Gene of interest
A specific gene that is identified and isolated for insertion into a plasmid during the cloning process.
Aqueous solution in sequencing
The medium in which DNA fragments are isolated with beads during the sequencing process.
Multiwell plate
A plate containing 2 million wells, each holding a different DNA fragment to be sequenced.
Pattern of flashes
The sequence of light signals generated during nucleotide addition, revealing the original DNA fragment's sequence.
Electric current interruption
The method used in nanopore sequencing to identify bases by their effect on an electric current.
Recombinant plasmid
A plasmid that has been modified to contain a gene of interest.
Recombinant bacterium
A bacterial cell that contains a recombinant plasmid.
Cloning vector
A plasmid used to clone a foreign gene.
Restriction enzyme
An enzyme that cuts DNA molecules at specific sequences called restriction sites.
Restriction fragments
The pieces of DNA that result from the cutting of DNA by restriction enzymes.
Sticky ends
Single-stranded ends of DNA fragments that can bond with complementary sticky ends of other fragments.
DNA ligase
An enzyme that seals the bonds between restriction fragments.
Bioremediation
The use of living organisms, such as bacteria, to remove or neutralize contaminants from a polluted area.
Human growth hormone
A protein produced by recombinant DNA technology to treat stunted growth.
Therapeutic proteins
Proteins produced through genetic engineering for medical treatments, such as those that dissolve blood clots.
EcoRI
A specific restriction enzyme that cuts DNA at the sequence GAATTC.
CTCGAG
A DNA sequence recognized by a restriction enzyme that cuts between bases A and G.
Bacterial plasmid
A small circular DNA molecule from bacteria that can replicate independently.
Base pairing of sticky ends
The process by which sticky ends of foreign DNA and plasmid DNA pair by complementary base pairing.
Sugar-phosphate backbone
The structural framework of nucleic acids, consisting of a chain of sugar and phosphate groups.
Recognition sites
Specific sequences in DNA where restriction enzymes cut.
Culture
A method of growing bacterial cells in a controlled environment.
Multiplication of recombinant bacterium
The process by which a recombinant bacterium divides and grows to form a clone of cells.
Pest-resistant crops
Genetically modified plants designed to resist pests.
Toxic waste cleanup
The process of using genetically modified bacteria to clean up environmental pollutants.
Foreign DNA segment
A DNA segment from another source that is inserted into a plasmid.
Agarose
A polysaccharide used to create the gel for gel electrophoresis.
DNA-binding Dye
A dye that fluoresces pink in UV light, used to visualize DNA bands after gel electrophoresis.
Denaturation
The first step in PCR where heat is used to separate double-stranded DNA into two single strands (at around 94-98°C).
Taq Polymerase
A heat-stable DNA polymerase used in PCR to synthesize new DNA strands.
Pfu Polymerase
A more accurate and stable DNA polymerase than Taq, used in PCR.
PCR Primers
Short single-stranded DNA molecules that are complementary to sequences flanking the target DNA region.
Cycle in PCR
Each cycle doubles the number of DNA molecules, typically resulting in millions to billions of copies after 20-35 cycles.
Restriction Sites
Specific sequences in DNA that can be cut by restriction enzymes, designed into PCR primers for cloning.
Preparation of DNA Fragments
The process of generating DNA fragments using PCR, designed to include restriction sites.
Cutting with Restriction Enzymes
The process where both the DNA fragment and the cloning plasmid are cut using the same restriction enzymes.
Electrophoresis Visualization
The process of visualizing separated DNA fragments as pink bands under UV light after gel electrophoresis.
Current in Gel Electrophoresis
The electrical current that causes negatively charged DNA molecules to move toward the positive electrode.
DNA Molecule Movement
Shorter DNA molecules move faster through the gel than longer ones during gel electrophoresis.
Amplified DNA Strands
DNA strands produced by PCR that may occasionally incorporate errors.
Recombinant DNA plasmid
A circular plasmid containing both the original bacterial DNA and the inserted foreign gene.
Antibiotic resistance gene
A gene carried by the cloning vector (plasmid) that allows bacteria to survive in the presence of antibiotics.
Successful transformers
Bacteria that have taken up the plasmid and can survive in an antibiotic medium.
PCR
A technique that amplifies DNA from various sources for biological research and genetic engineering.
cDNA
Complementary DNA that contains only exons, used to avoid problems with introns in eukaryotic genes.
Expression vector
A cloning vector that contains a highly active bacterial promoter to facilitate gene expression.
Eukaryotic cells
Cells that can be used as hosts for cloning and expressing genes, avoiding incompatibility issues with bacterial systems.
Electroporation
A method of introducing recombinant DNA into eukaryotic cells by applying a brief electrical pulse.