Bio 106: Transcription & Translocation
1/71
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
72 Terms
chromosome
structure that consists of DNA and associated proteins, carries part (eukaryotes) or all (bacteria) of a cell's genetic info for making new organism
DNA in eukaryotic cell __ is organized as one or more chromosomes that differ in length and shape
nucleus (humans have 22 pairs of autosomes, 1 pair of sex chromosomes)
gene
A segment of DNA on a chromosome that codes for a specific trait; encodes a protein (functional production)
transcription and translocation
expression of info in a gene occurs in these 2 stages
Central Dogma of Molecular Biology (Watson and Crick)
DNA contains original codes for making proteins that living things need; DNA --(transcription)--> RNA --(translation)--> protein
mRNA
messenger RNA; type of RNA that carries instructions from DNA in the nucleus to the ribosome; ribosome then reads its coding sequences and puts the appropriate amino acids together
transcription
process of converting a nucleotide sequence of a gene (genetic info in DNA) into a complementary strand of RNA; produces an RNA copy (TRANSCRIPT!) of a gene
complementary strand of RNA in transcription is made by
RNA polymerase: messenger RNA (mRNA), a single stranded RNA (ssRNA) molecule
RNA polymerase vs DNA polymerase
RNA polymerase uses DNA as a template to make RNA; DNA polymerases uses DNA as a template to make DNA
both are DNA-dependent
RNA contains __ in place of thymine
uracil (A-U and G-C base pairings between DNA and RNA)
RNA uses __ in place of deoxyribose nucelotides
ribose nucleotides
oxygen (hydroxyll group) in RNA explains why
RNA lasts very SHORT time, is catalytic, but UNSTABLE in comparison to DNA that is missing oxygen and lasts very long time
3 stages of transcription
initiation, elongation, termination
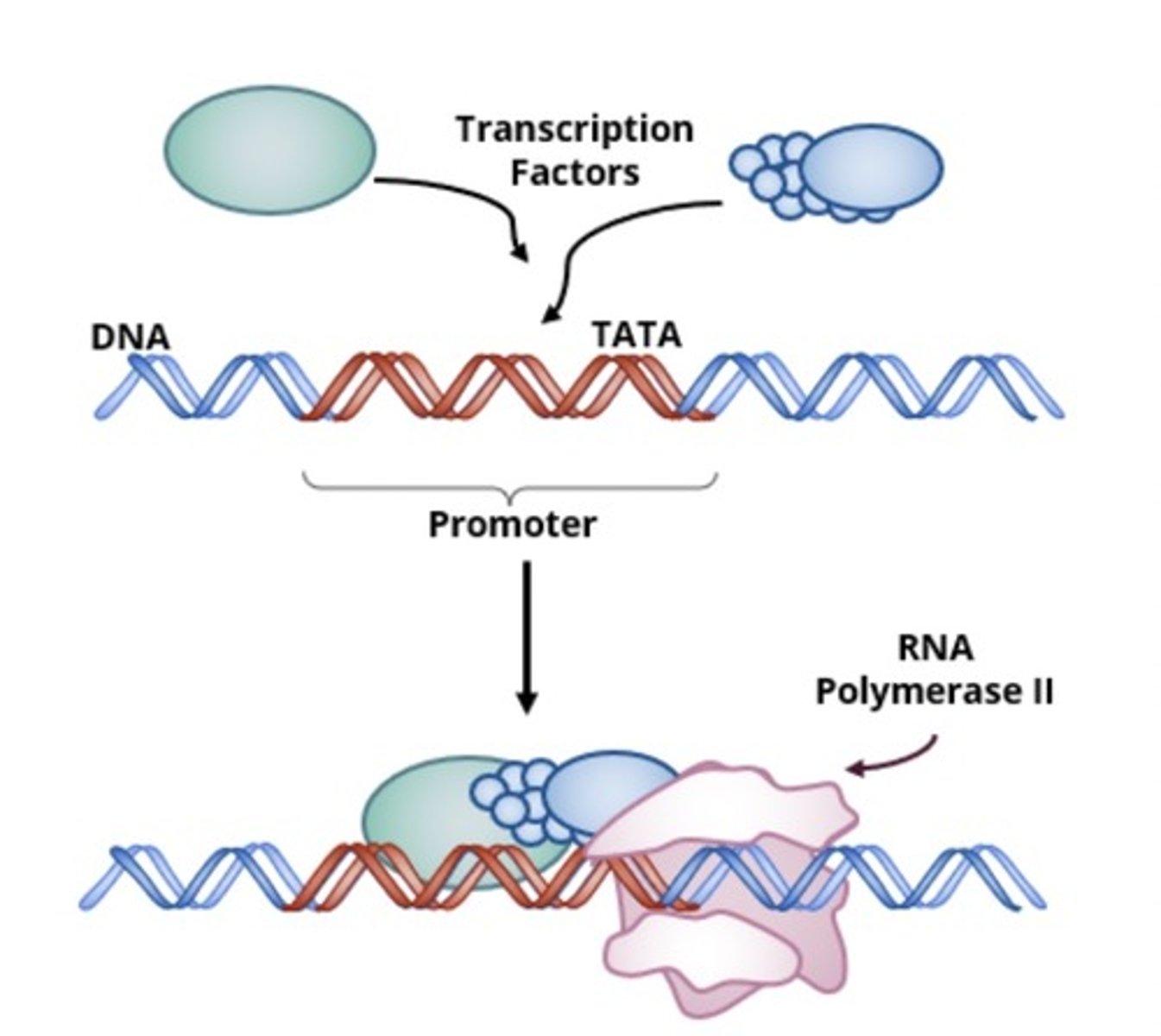
initiation (transcription)
recognition step; RNA polymerase and regulatory proteins bind to promoter (most time-consuming, complicated step in transcription)
elongation (transcription)
RNA polymerase synthesizes RNA, linking nucleotides together (5' to 3')
termination (transcription)
termination sequence causes polymerase and RNA transcript to dissociate from DNA
role of promoter in transcribing a gene into RNA
signaling the beginning of transcription; sequence of DNA where polymerase binds
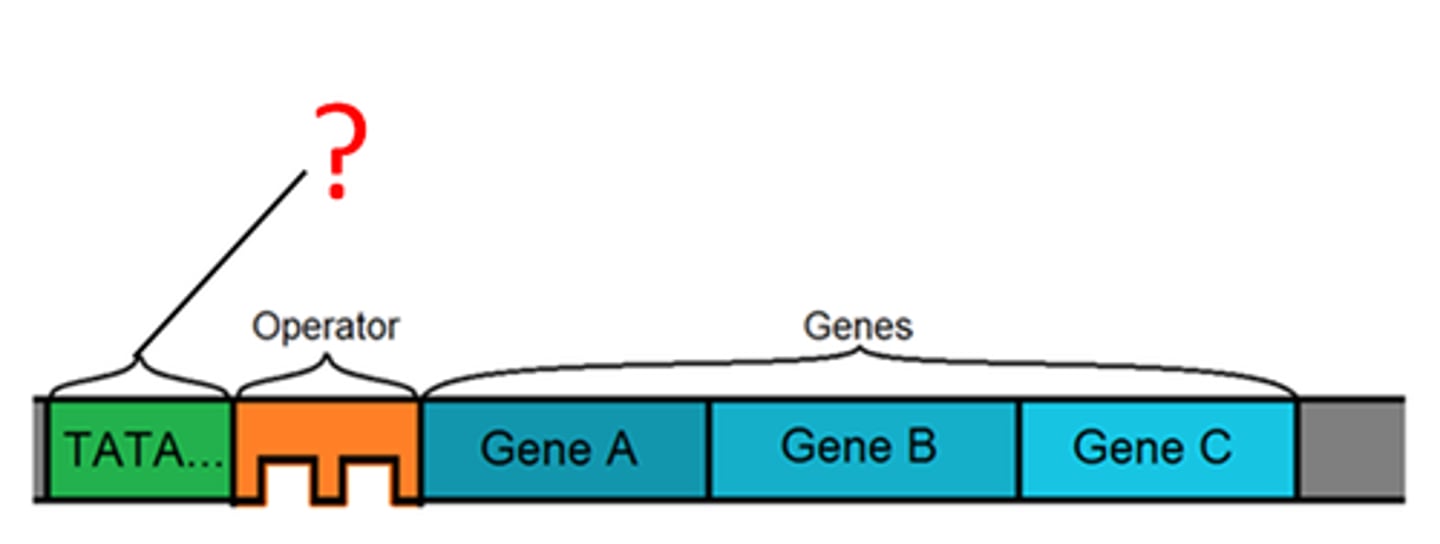
role of regulatory sequence in transcribing a gene into RNA
site for binding of regulatory proteins; influences the rate of transcription -- determines what and when genes are expressed
transcribed region
contains the information that specifies an amino acid sequence for mRNA
terminator region on DNA
signals the end of transcription
transcription initiation factors
specialized proteins that assist in formation of pre-intiation complex; RNA polymerase requires these factors to LOCATE and BIND TO promoter regions of genes (different factors dependent on gene type)
enhancer in DNA
Regions of DNA where factors that regulate transcription can also bind
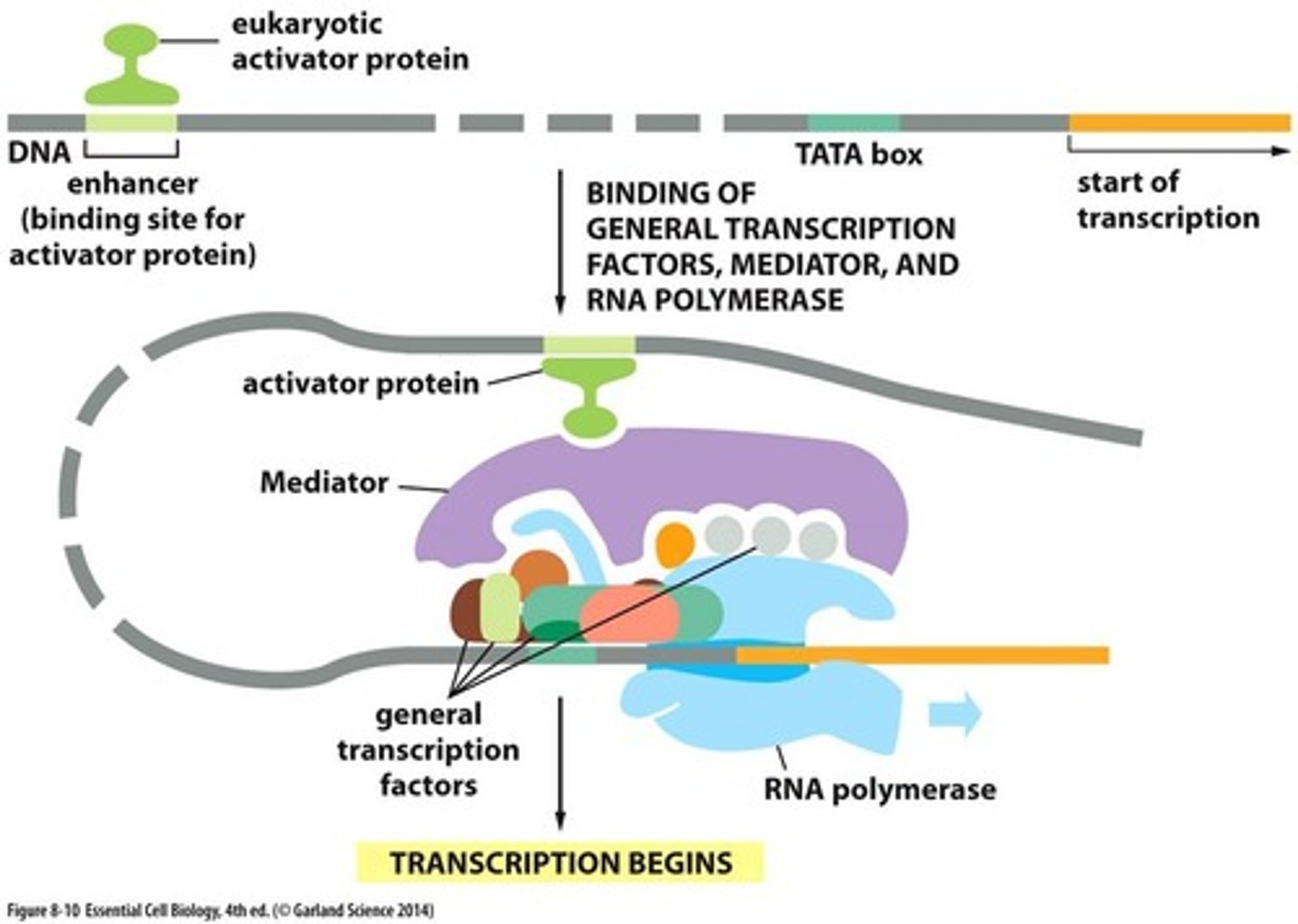
activator and mediator proteins
link bridge between enhancer and transcription factors
TATA box
A promoter DNA sequence crucial in forming the transcription initiation complex.
sigma factors
proteins in prokaryotic (BACTERIA) cells that bind to RNA polymerase and direct it to specific classes of promoters; assists RNA pol. to find and bind promoter
polymerase holoenzyme
polymerase with other factors bound
in DNA replication, helicase is needed to unzip DNA; in RNA replication during initiation step, __ acts as its own helicase (doesn't matter if DNA closes back up)
RNA polymerase
during elongation, copy of RNA is made from __ strand
template (antisense)
RNA synthesis occurs in __ direction
5' to 3'
RNA sequence made during elongation is equivalent to __ (or coding strand)
non-template (sense)
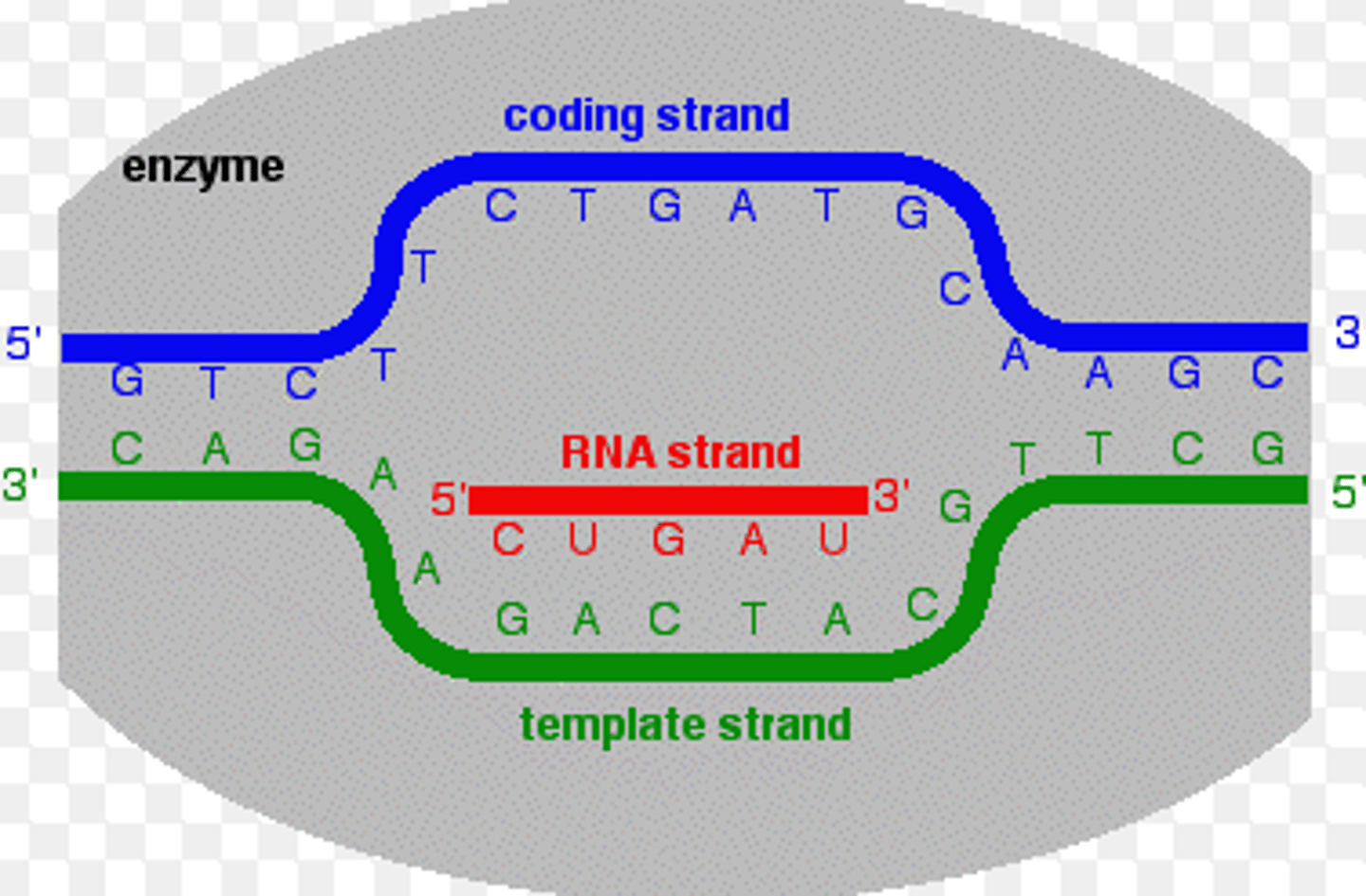
__ strand of DNA can be used as template in elongation
either
termination stage of transcription =
not well understood; different for different systems; often involves looping that prevents DNA polymerase from getting through
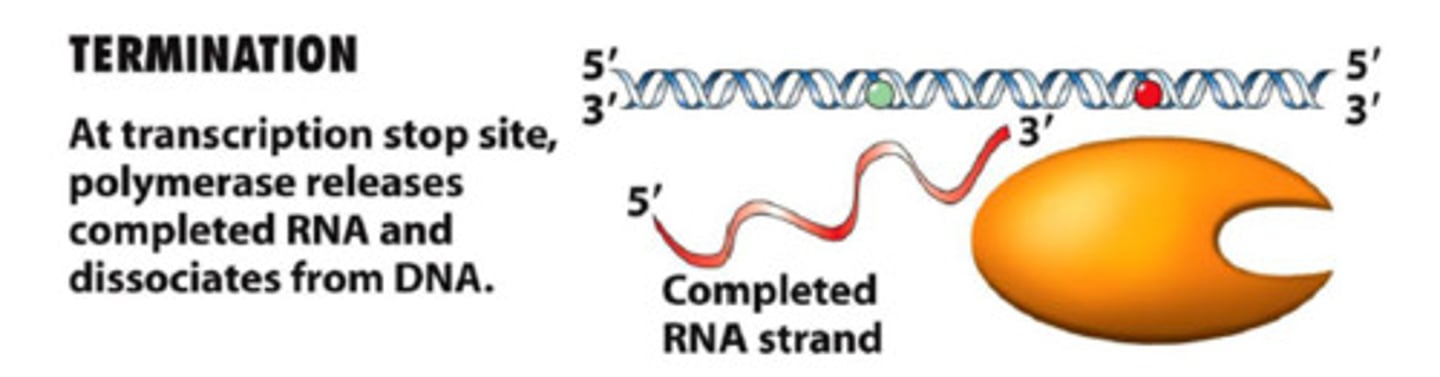
one gene can be transcribed by
multiple RNA polymerases all at once
RNA transcription occurs in nucleus (in eukaryotes) but info is converted into protein in __
cytoplasm
post-transcriptional modifications in eukaryotes
changes in RNA before leaving the nucleus in eukaryotes
pre mRNA consists of
introns and extrons
introns
nucleotide sequences that are removed from a new RNA; snipped OUT
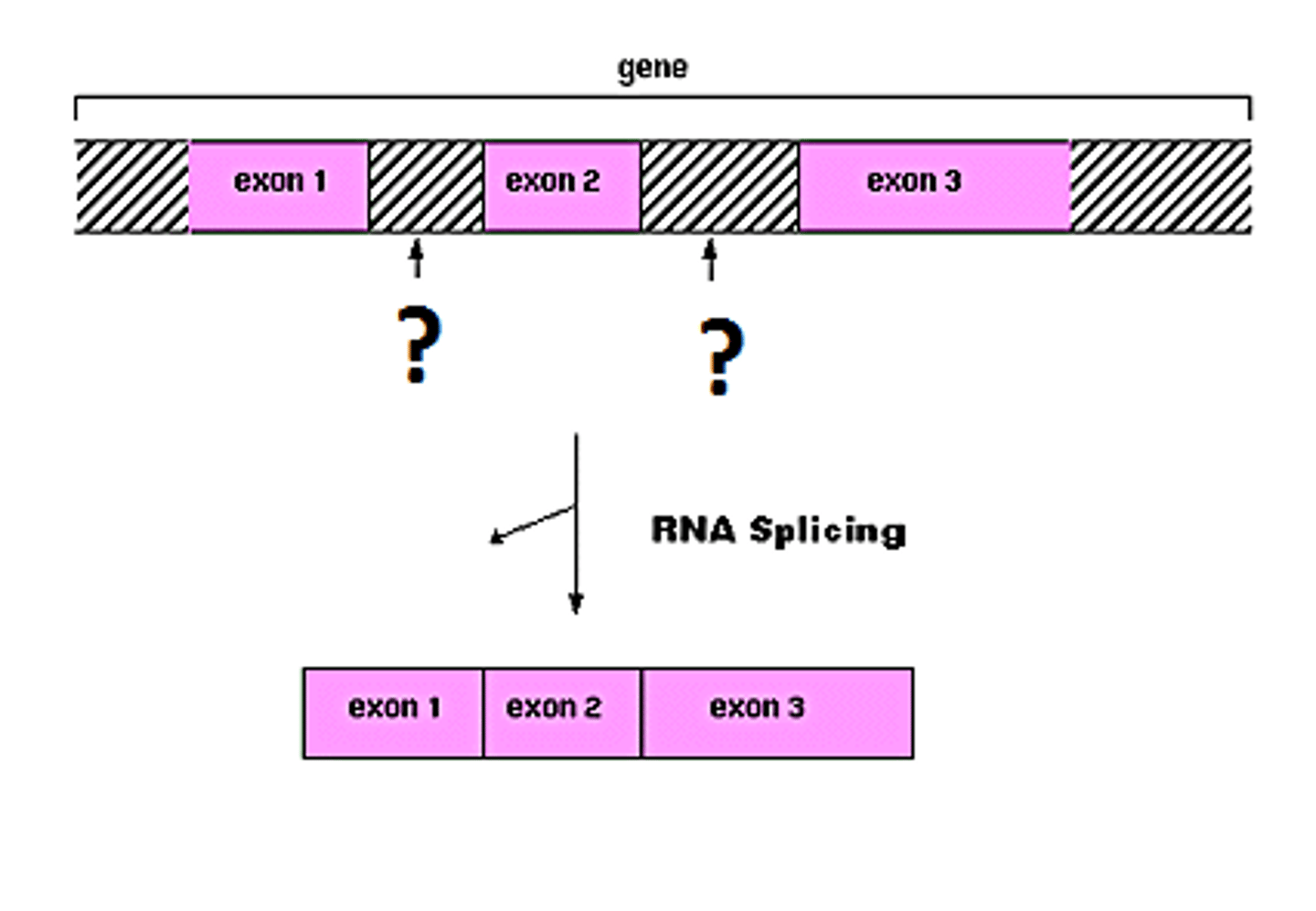
Extrons
Sequences of RNA that are translated into proteins; stay IN the RNA
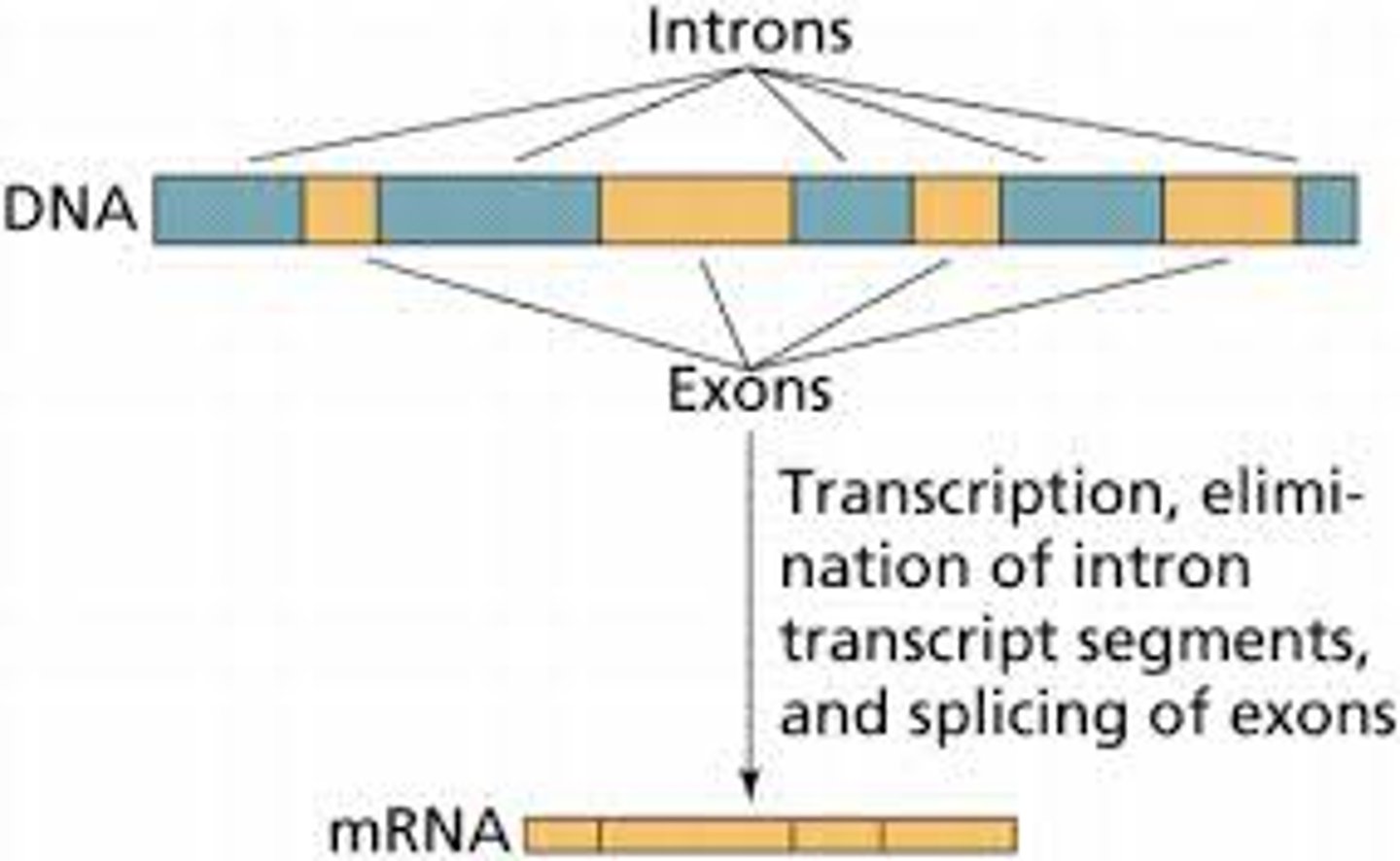
mature mRNA contains only
exons
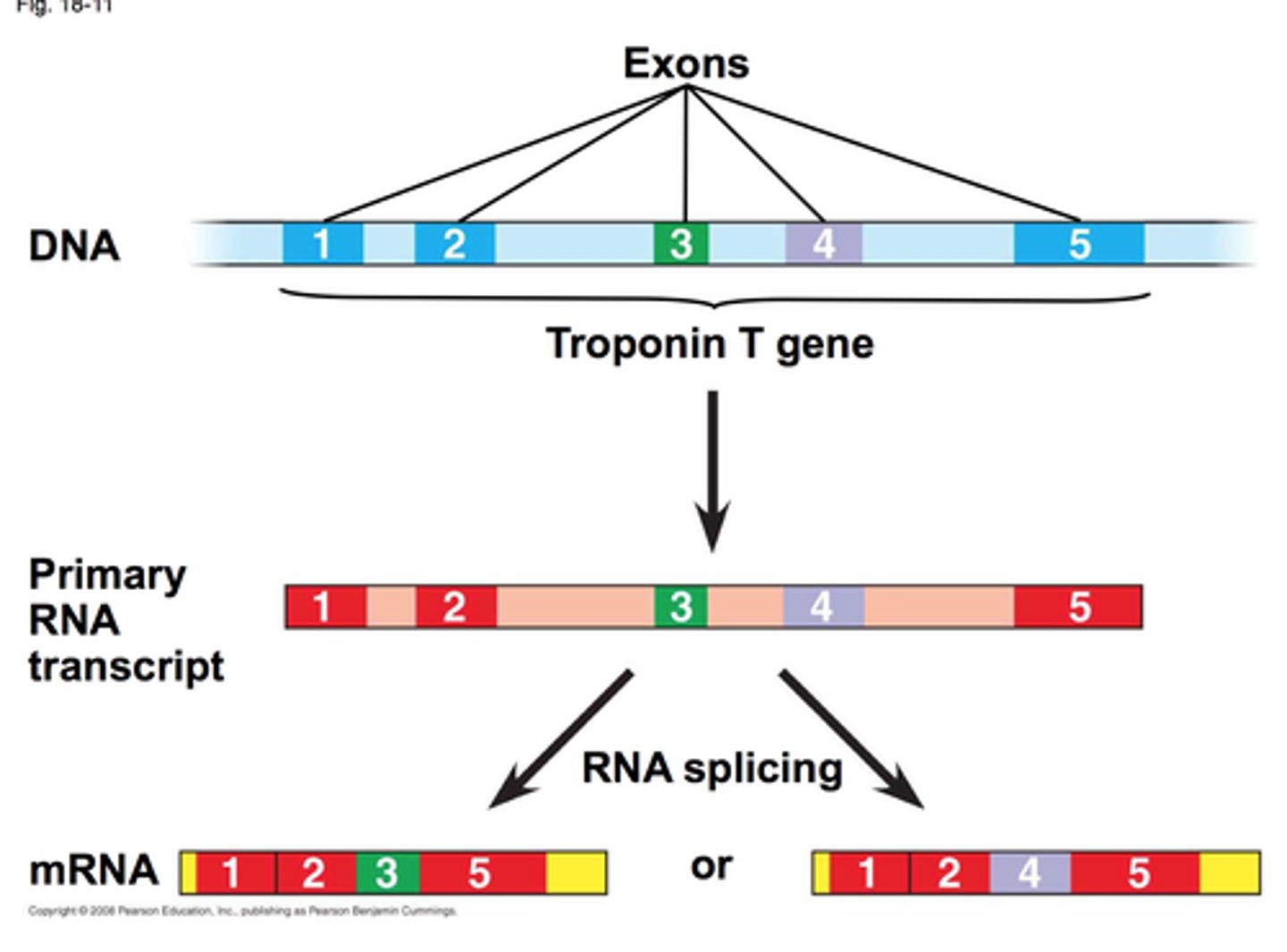
alternative splicing
allows a single gene to encode multiple proteins; can piece together different exons that result in different proteins being made; allows cells to increase coding capacity without increasing amount of DNA (viruses do this best!)
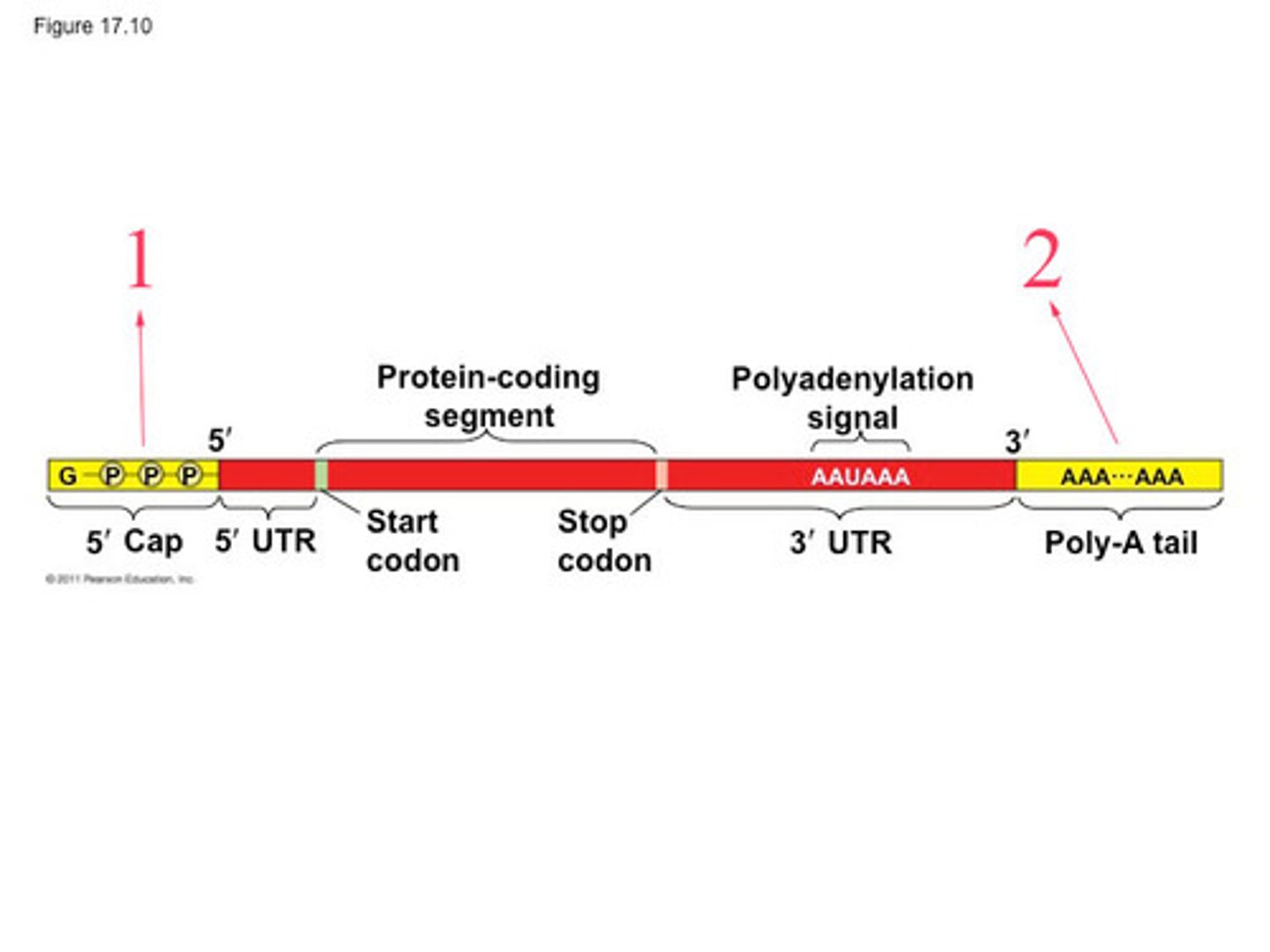
mRNA 5' caps play a role in
RNA stability, nuclear export, translation; cap = 5' carbon on guanine
Poly(A) tails
added after transcription
- enhance mRNA stability, nuclear export, translation efficiency
- play a role in transporting the mature mRNA from the nucleus to the cytoplasm
similar to caps
sequence that tells enzymes to cleave
translation
information carried by mRNA is decoded into linear sequence of amino acids, resulting in polypeptie (protein) chain that folds into protein
protein translation occurs on
ribosomes
Central Dogma (flow of genetic information)
DNA -> RNA -> Protein
4 basic units in DNA/RNA (adenine, cytosine, guanine, thymine/uracil) are converted into __ basic units (amino acids) found in proteins
20
features of genetic code
64 different 3 nucleotide "codons"; nonoverlapping; 3 different "reading frames" on each strand; 6 possible reading frames; redundancy of genetic code; only 20 amino acids to be encoded by 64 possible codons in the triplet code; most amino acids specified by two or more codons; synonymous codons specify same amino acid
linear order of codons in mRNA determines
linear order of amino acids in a polypeptide chain
codon
sequence of THREE mRNA nucleotides that codes (specifies) for a specific amino acid; reading in 3 basic chunks creates reading frames
length of a codon is 3 because
length needed to give at least 20 unique codons (corresponding to 20 unique proteins)
which codon specifies for methionine (met)?
AUG
starting codon
AUG (methionine)
some amino acids can be coded by
more than 1 codon (most have 2-3 variations)
mRNA is read from
5' to 3' by ribosome
stop codons (no amino acid specified to them)
UAA, UAG, UGA
not typical for a protein/ribosome to start reading at exact start of a sequence because
this can cut the sequence short
to determine which strand in double helix DNA is the template for mRNA
look for AUG start codon
Mis-sense substitution mutation
amino acids have been changed
silent mutation
A mutation that changes a single nucleotide, but does not change the amino acid created
nonsense mutation (truncation)
insertion of stop codons "knock out" a gene to stop proteins from being made --> results in protein being too short
frameshift mutation
mutation that shifts the "reading" frame of the genetic message by inserting or deleting a nucleotide --> can result in protein being too short/long
translation machinery consists of
mRNA (messanger RNA)
ribosomes, which link amino acids into polypeptide chains
- each ribosome consists of 2 subunits - 1 large and 1 small
- both subunits contain rRNA (ribosomal RNA) and proteins
ribosome structure (general structure same for prokaryotic and eukaryotic ribosomes)
large subunit,small subunit
translation involves 3 types of RNA
messenger RNA, ribosomal RNA, transfer RNA
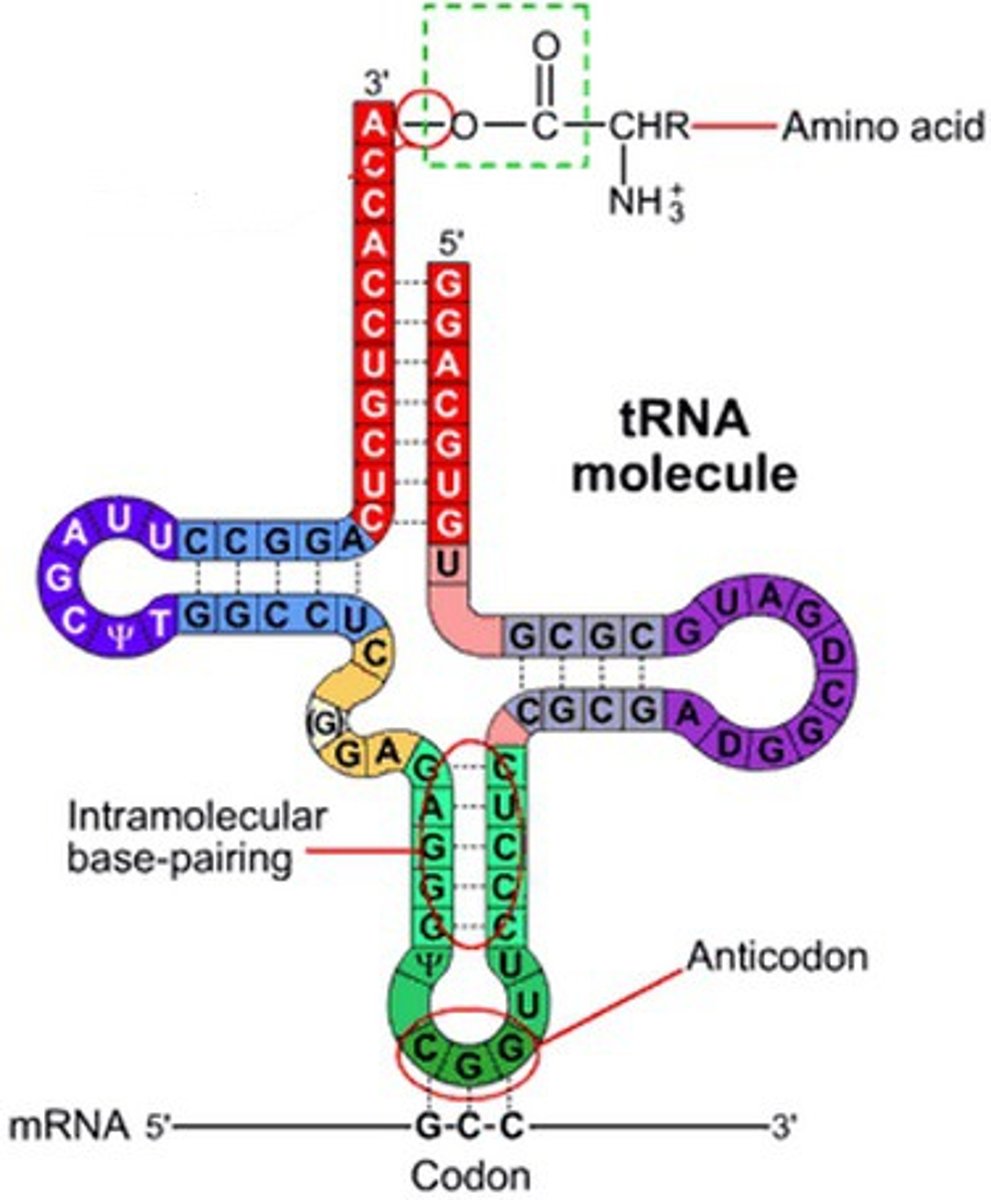
transfer RNA
type of RNA molecule that delivers amino acids to ribsomes; has an ANTICODON that is complementary to an mRNA codon
tRNA structure
anticodon at one end; codon-anticodon connection is complementary and antiparallel
amino acid attachment site at other end
3 stages of translation
initiation, elongation, termination
initiation (translation)
1- initiation complex is formed when small ribosomal subunit binds to mRNA
2- anticodon of intiator tRNA base-pairs with the start codon (AUG / Met) of mRNA
3- large ribosomal subunit joins small ribosomal subunit
intitation complex (small + large ribosomal subunit) consists of
3 sites: E,P,A
elongation (translation)
ribsome assembles polypeptide chain as it moves along mRNA; initator tRNA carries methionine (first amino acid in chain) into P site
(lagre subunit) ribosome joins each amino acid to polypeptide chain with a peptide bond
order of tRNA movement during elongation (translation)
A site --> P site --> E site ("ejector site")
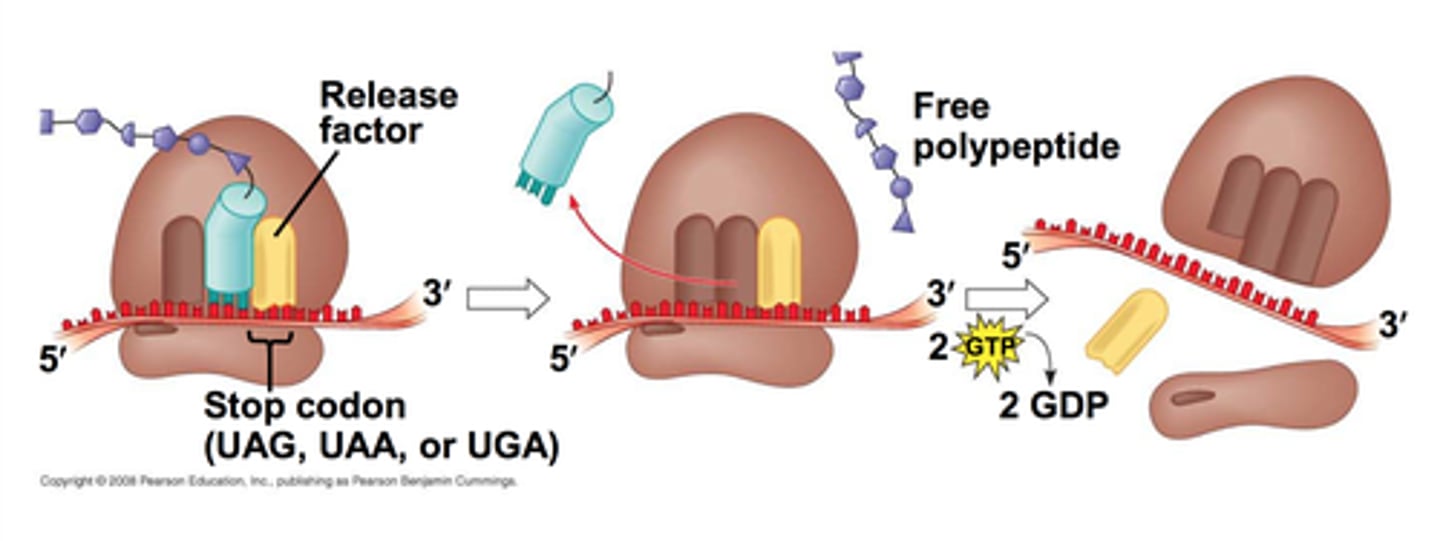
termination (translation)
when ribosome encounters a stop codon, polypeptide synthesis ends
- release factors bind to ribsome
- enzymes detach the mRNA and polypeptide chain from ribosome
stop codons cause everything to fall apart (still sequence that won't be translated)