A3.2 Classification & Cladistics
1/24
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
25 Terms
How do we classify organisms? What is the definition of Classification for living things?
We use a hierarchical system; So we start really broadly, and then get more specific as we move down a list of criteria. Classification is sorting and organizing knowledge of living things based on their characteristics, and most recently their DNA.
What is the definition of Taxonomy?
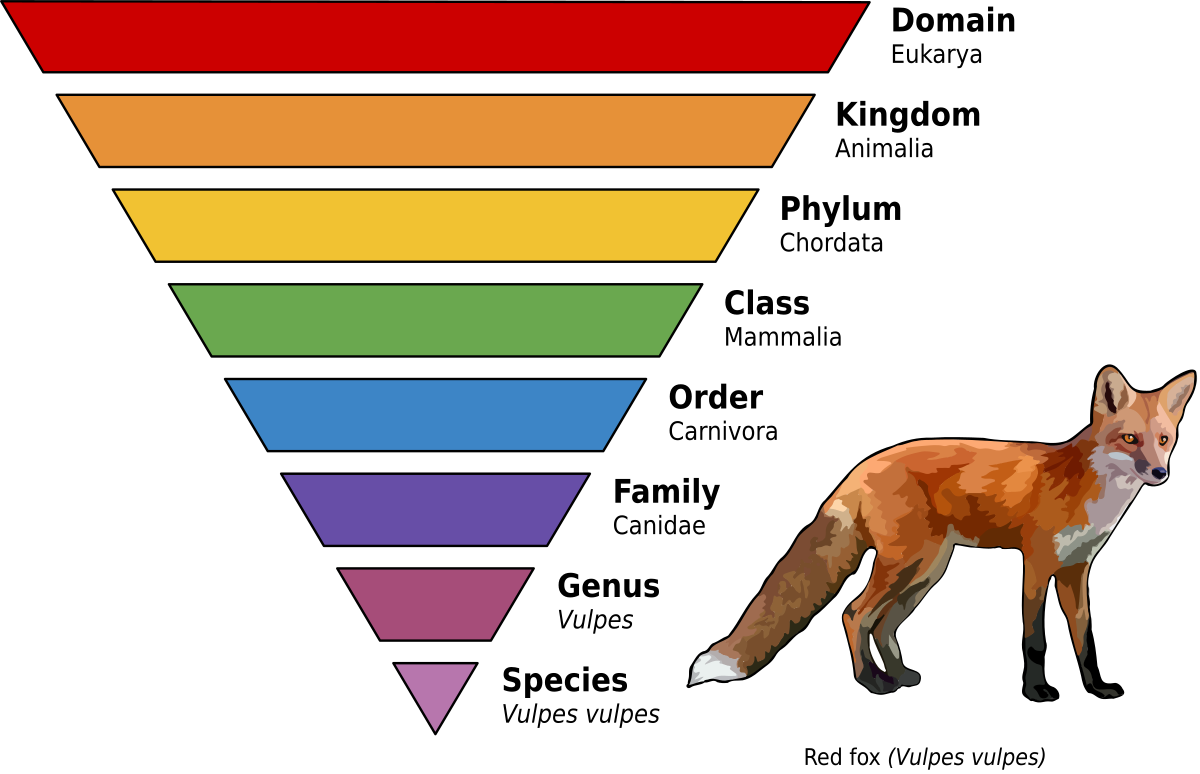
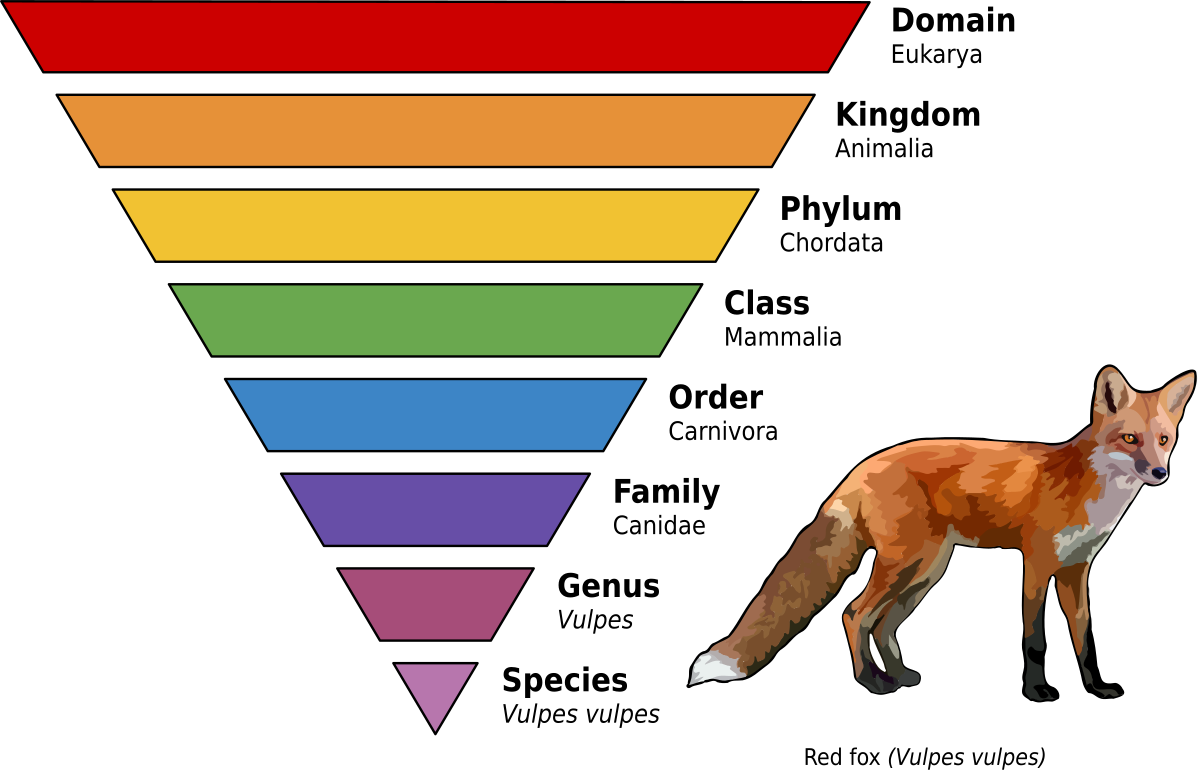
The process of assigning an organism to a particular group (known as a taxa). Domain is the broadest level of grouping (and it includes: archaea, bacteria, and eukarya/eukaryotes), while Species is the narrowest level of grouping.
What are some of the advantages of having a classification system?
Classification systems allow us to:
Better Organize & Understand the world by grouping similar things
Better communicating ideas by creating a common language to refer to certain groups of things
Quickly & Easily find and categorize information
Conduct further research to identify trends and patterns in the things we’ve classified
Who is Carl Linnaeus, and what is it that he did?
Carl Linneaus was a swedish scientist who created the classification system we still use today. The classification system he created is called the: “Hierarchy of Taxonomy” He’s known as the father of taxonomy. His classification system was based on shared characteristics, which we still use to classify organisms today, although we have adjusted and added to his original hierarchy of taxonomy.
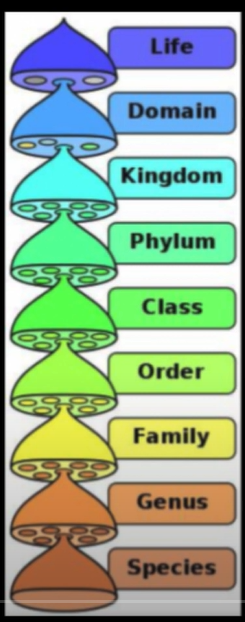
What parts make up the Hierarchy of Taxonomy? What are intermediate taxa?
DOMAIN
KINGDOM
PHYLUM
CLASS
ORDER
FAMILY
GENUS
SPECIES
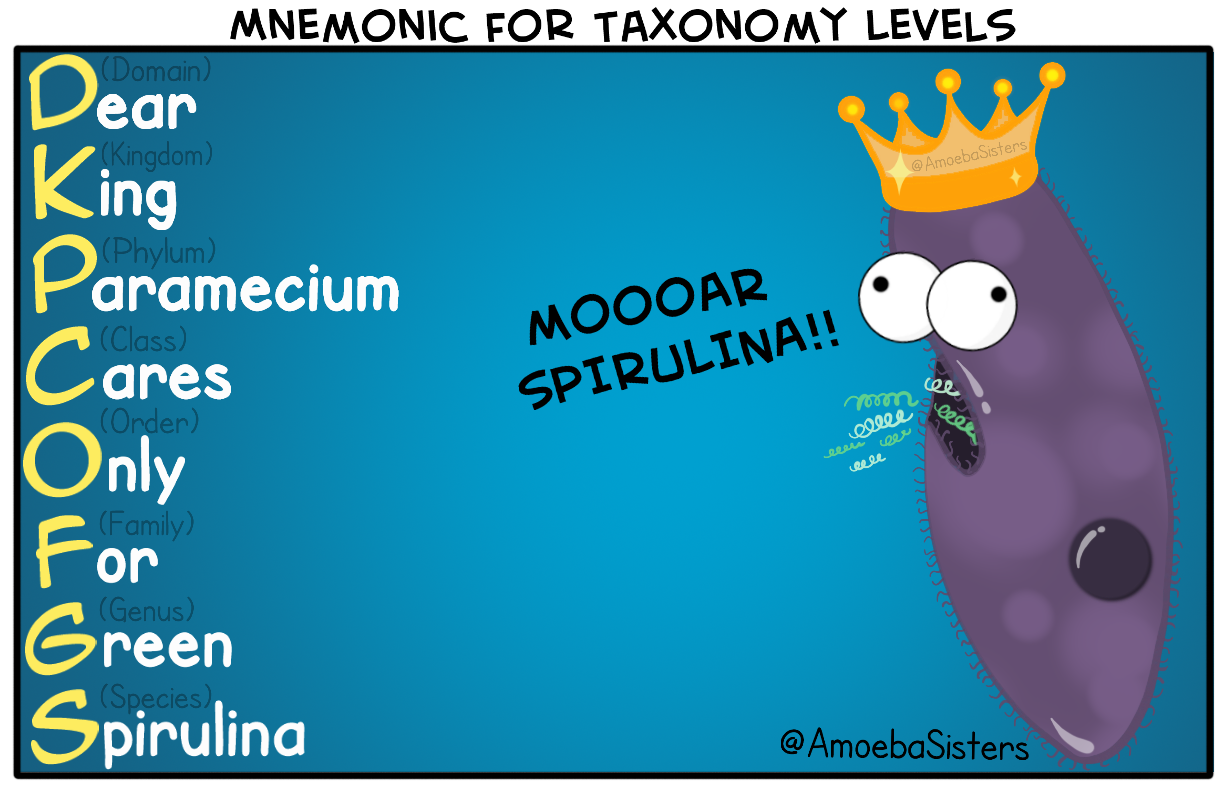
Dear King Paramecium Cares Only For Green Spirulina
Intermediate Taxa are subfamilies that have been added in between these Taxa.
What’s the problem with the traditional Hierarchy of Taxonomy?
IT IS ARBITRARY (YOU MUST USE THIS WORD TO GET THE FULL MARK), because it is based on observable features which are subjective, and don’t always show the complex evolutionary patterns organisms have gone through over billions of years;
The classifications were initially only based on shared characteristics (similar features, a similar environment) and didn’t take into account genetics at all. But after more recent developments, we’ve figured out that some organisms needed to be reclassified
Give an example of one organism that needed to be reclassified to a different species due to new discoveries in genetics?
The giant panda and red panda were re-classified; Originally we thought that red pandas and giant pandas were part of the same species because they had similar features, a similar diet and habitat; But after a genetic analysis, they found that giant pandas are actually members of the bear family, and red panadas are more related to racoons, weasels and skunks.

What are the advantages of re-classifying things based on new genetic data?
It shows the evolutionary relationships between different organisms, which can help us group them based on common ancestors
It can better reflect the process of evolution
It can be used to make predictions about the characteristics of organisms
It can be used to understand the diversity of life on earth
In the hierarchy of Taxa, what is in a family?
The taxon found directly below family is genus, therefore, a family contains genera.
Organisms A, B and C belong to the same class but to different orders.
Organisms D, E and F belong to the same order but to different families.
Which of the following pairs of organisms would be expected to be the most similar?
Class is a higher taxon, and it’s made up of orders. So if organisms belong to an order, then this means that we’ve already narrowed it down a bit more from Class, and so they share more characteristics. This means that organisms D, E, and F are most similar.
What is a clade? What is a key feature of Clades? What are cladistics? How do we figure out evolutionary history?
Clade = A group of species with a single common ancestor & a set of common traits/characteristics
Clades are hierarchical, meaning they can be divided into smaller subclasses/subsections; For example, Mammals are made up of the subclass Prototheria, and then the subclass Theria, and so on and so forth.
Cladistics = A way of identifying evolutionary relationships between species based on shared traits & genetic evidence
We can figure out the evolutionary history of different species by comparing their molecular sequences (DNA sequences) or physical traits.
What type of unbiased evidence tells us that two species belong to the same clade? Give an example of two different species that we’ve identified to be more similar to each other than we thought before.
Why are physical traits a less reliable method of classifying species into clades?
Molecular sequences gives us the most unbiased evidence. By comparing the base sequences of a gene on chimps and humans, we’ve figured out that chimps are more similar to humans than to gorillas.
Physical traits are less reliable because of things like convergent evolution (unrelated species evolving to have similar characteristics), and mimicry (when a species evolves to have characteristics that resemble a different organism in order to give them a certain advantage).
Define Cladistic analysis? What is the idea of cladistic analysis based on?
The technique of classifying organisms into clades based on their shared characteristics and most recent common ancestor. Cladistic analysis is based on the idea that shared genes or traits have been passed down from a common ancestor and define a clade.
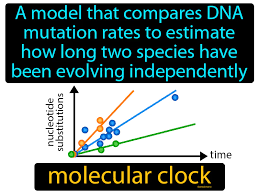
What is the molecular clock? Why does this method work? What is a calibration point?
The molecular clock measures the rate at which the gene mutations of a species accumulate in order to determine how long ago two species shared a common ancestor.
A calibration point is a known, precise moment when two species diverged, to help scientists calculate the divergence time of other species in reference to this time. A calibration point can be in the form of the oldest fossil record of a specific species.
This method works because genetic mutations increase over time as a result of evolution.
If a particular mitochondrial gene accumulates base sequence mutations at a rate of 3 every 5 million years, about how long ago would two species have shared a common ancestor if their gene base sequence differs by 13 bases?
We know that 3 mutations occur every 5 million years. This means that 1 mutation occurs every 1.67 million years.
So if 13 base sequence mutations have occurred, then this means that it has been
13/1.67 million years = 21.7 million years since the two species would have shared a common ancestor.

What are cladograms? How are they made?
Cladograms are diagrams that visually represent the evolutionary relationships between different groups of organisms; Cladograms show their relationships based on shared characteristics and common ancestors.
Cladograms are made using cladistic analysis (so that can include both shared physical and genetic traits); First scientists choose a specific species that they want to compare, then they pick traits (physical features, behaviors or genetic traits) that they want to compare; For example: comparing a bat, bird and a ladybug for the traits of wings (present or absent?), feathers (present or absent?), fur (present or absent?). Afterwards, a matrix is made, where the species are listed on one axis, and the traits are listed on the other axis. The matrix shows which species have which traits; Then this matrix is used to create a cladogram.
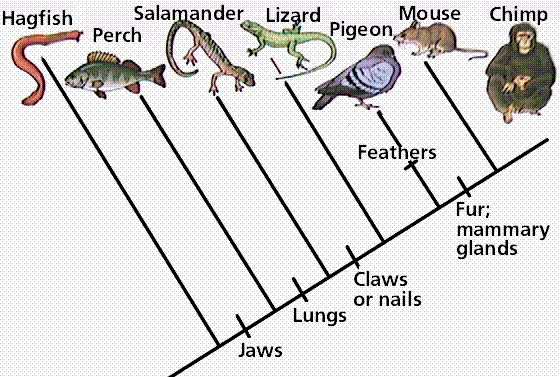
How are species grouped on a cladogram?
On a cladogram, scientists try to group together species with similar traits, because they’re more closely related to each other. They also try to place species in a way that makes most sense based on the way they evolved and create a tree with the fewest number of sequence changes or trait differences. For example, it makes more sense to put egg-laying mammals like the playtpus in the mammal group, because putting them there requires fewer developments of new characteristics and fewer changes in traits then putting them in a totally different group like the reptile group (even though they also lay eggs)
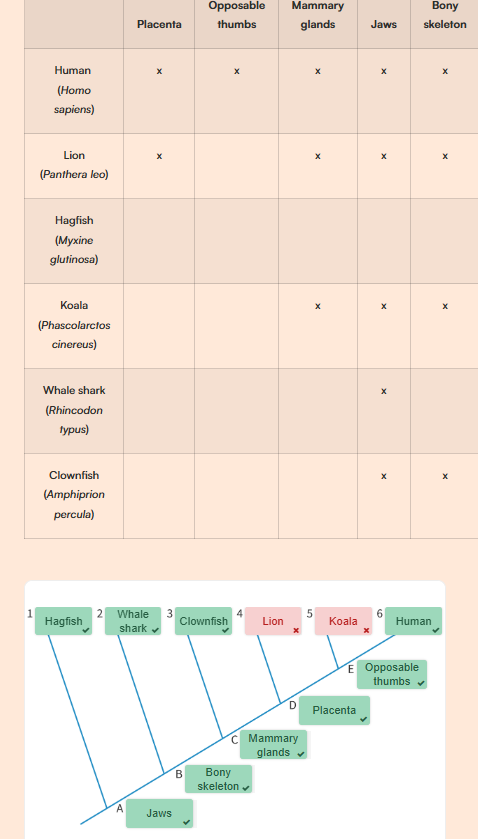
Label the Cladogram below. What are the three key terms you need to know?
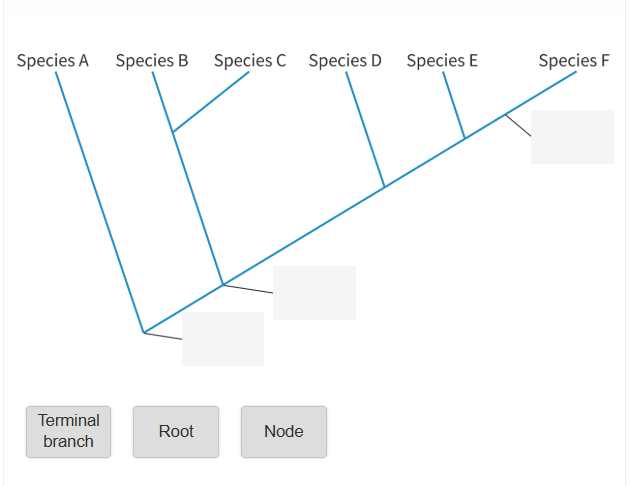
Answer to the Cladogram labelling:
Root = the most ancient common ancestor
Node = the hypothetical common ancestor
Terminal Branch = the end branches of a cladogram, representing a still existing species.
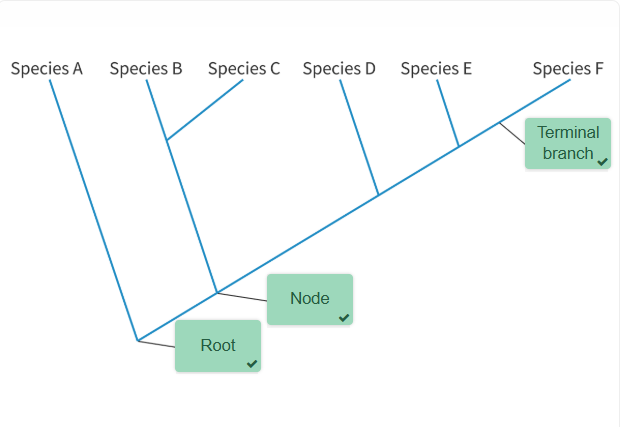
What are some of the assumptions that a cladogram is based on (think: What does the branching pattern represent on a cladogram? What needs to be explicitly mentioned if extinct species are included on a cladogram? What does having more nodes in between species mean?)
The branching pattern represents the evolutionary relationship between species.
If extinct species are included, scientists have to be disclose whether the cladogram is based on morphology only; since there is usually no DNA data available for this type of analysis.
Having more nodes in between species means that their relationship is more distant.
We assume that mutations at the DNA or protein level happen at a constant rate; for example: one base change every 10^9 (billion years)
We assume that some cladograms are drawn to scale, and that the branches accurately represent the time since divergence.
Which two species are most closely related and which are least related on this diagram?
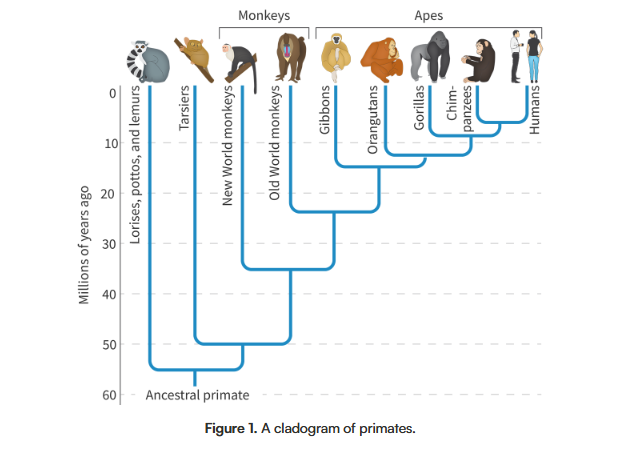
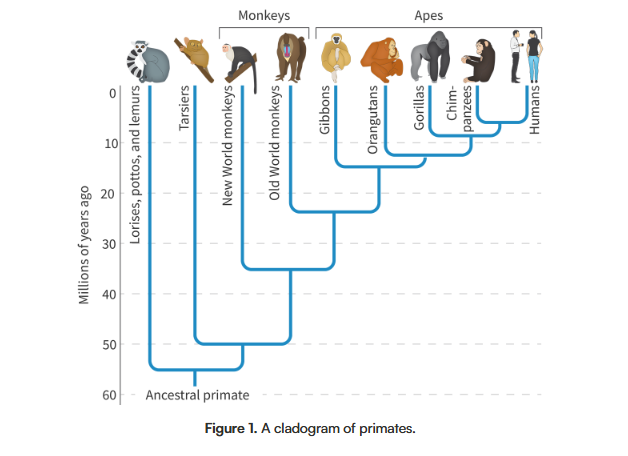
Answer to the previous question:
Humans and chimpanzees are most closely related, while lemurs are the least related to anything else on the graph.
What can we estimate from a cladogram? What is this conclusion being compared to?
We can estimate when a species diverged, and when their common ancestor existed. After analyzing the molecular differences of different animals and comparing them to the oldest fossilized records, for example, we can conclude from a cladogram that humans and chimps shared a common ancestor about 6-7 million years ago.
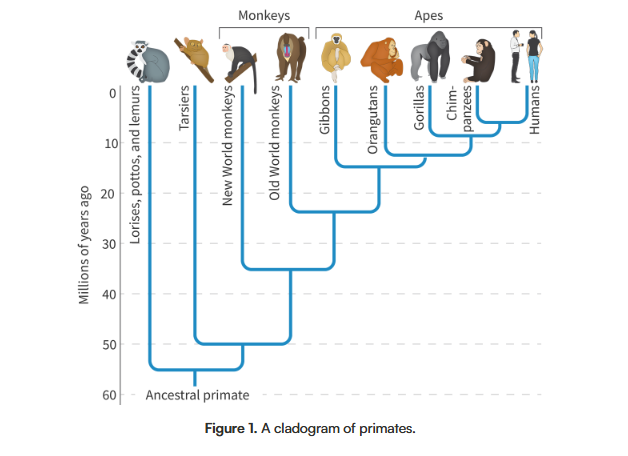
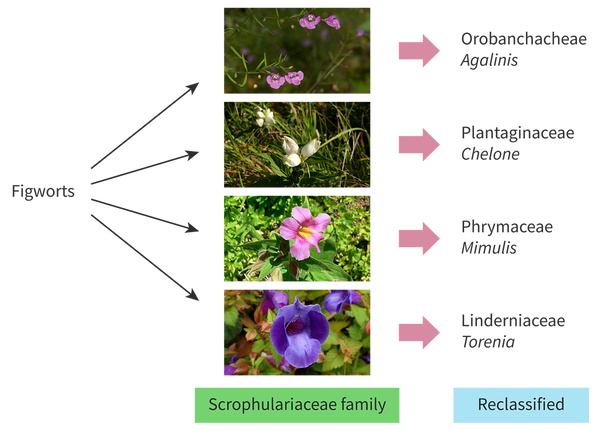
Give an example of one family of plants that was reclassified as a result of us adding molecular data to cladograms
The figwort family (family of flowers) was reclassified after an analysis was conducted on a gene found in their chloroplasts. Before, they included more than 275 genera (genuses) with 5000 species, but after this genetic analysis, they found that many species within this group were actually very different from each other, and did not all come from the same common ancestor. Now instead of these flowers all belonging to a single clade, they make up five separate clades; So the Figwort family has only 200 different species now. Only half of the plants that were originally classified as figworts are still a part of the figwort family.
Who was Carl Woese, and what did he suggest? Why did he suggest this? Why was it important? (not important)
Carl Woese was a microbiologist, and in 1977, he suggested that we add a new taxonomic level above kingdoms: Domains. He suggested this after finding that RNA in ribosomes (otherwise known as rRNA sequences) formed three different/distinct groups: eubacteria, archaea, and eukarya. It also showed that eukaryotes were more closely related to archaea than to eubacteria.
His discovery completely changed the way we classified organisms and showed that molecular traits could accurately show the evolutionary relationships between different organisms.