CHEM314 FINAL FRQs
1/26
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
27 Terms
What is the approximate length of a DNA molecule (in the B form containing 1000 bases)
3400 A (3.4 A * 1,000 bp)
What repair systems exist, When There Is No Undamaged DNA to Use as a Template for Repair?
Homologous repair, Nonhomologous end loining (NHEJ), translesion synthesis (Error-prone)
What are the 4 steps in Homologous RecombinaIon?
1, 5’-end processing.
2, Strand invasion.
3, Branch migraIon.
4, Holliday intermediate resoluIon and ligaIon.
What is the key difference between HR and NHEJ?
HR involve used another Chromosome to synthesis DNA, but NHEJ does not have this step
To Amplify a Double-stranded DNA (a) in addition to the primers what additional materials
would be needed? (b) Design two 6-mer primers that could allowed you to amplify the following
DNA between the two symbols (< and >; write the primer sequences from 5’ to 3’) .
(5')ACTTCGGA<TCGTTA................................................AGGCCGCTT>TCTGT(3')
(3')TGAAGCCT<AGCAAT........................................................TCCGGCGAA>AGACA(5')
(a) Thermo-stable DNA polymerase, and dNTPs (dATP, dTTP, dGTP, dCTP)
(b) 5’TCGTTA3’, 5’AAGCGG3’
Mouse DNA hybridizes more extensively with human DNA than with yeast DNA.
Explain by describing the factor or factors that determine extent of hybridization.
In general, the more similar the sequences in two DNA molecules are, the more readily they will hybridize. Because the evolutionary distance between mouse and yeast is greater than that between mouse and human, mouse and human DNA sequences are more similar than those of mouse and yeast
Describe the composition and structure of a nucleosome.
A nucleosome consists of 146 base pairs of double-stranded DNA wrapped in a solenoidal supercoil around a core of histones (small, basic proteins) and 52 base pairs of linking DNA. This core contains two copies each of histones H2A, H2B, H3, and H4.
Briefly describe the biochemical role of the following enzymes in DNA replication in E. coli: (a) DNA helicase (b) primase (c) the 3' --> 5' exonuclease activity of DNA polymerase (d) DNA 1igase; (e) topoisomerases (f) the 5'-->3' exonuclease activity of DNA polymerase I
(a) Helicase unwinds double-stranded DNA during replication
(b) Primase synthesizes short RNA primers during lagging strand replication.
(c) The 3' --> 5' exonuclease activity of DNA polymerase proofreads newly synthesized DNA, removing mismatched nucleotides.
(d) DNA ligase seals nicks in the DNA at the boundaries between Okazaki fragments.
(e) Topoisomerases relieves the topological stress produced by the unwinding of double-stranded DNA at the replication fork.
(f) The 5' --> 3' exonuclease activity of DNA polymerase I removes RNA primers.
What defermines (in general) how much DNA will hybridize with another animals DNA?
In general, the more similar the sequences in two DNA molecules are, the more readily they will hybridize
DNA replication in E. coli begins at a site in the DNA called the (a) ___________. At the replication fork the (b) ___________ strand is synthesized continuously while the (c) _________ strand is synthesized discontinuously. On the strand synthesized discontinuously, the short pieces are called (d) ____________ fragments. An RNA primer for each of the fragments is synthesized by an enzyme called (e) __________, and this RNA primer is removed after the fragment is synthesized by the enzyme (f) ___________, using its (g) _____________ activity. The nicks left behind in this process are sealed by the enzyme (h) _____________.
(a) origin; (b) leading; (c) lagging; (d) Okazaki; (e) primase; (f) DNA pol I; (g) 5' to 3' exonuclease; (h) DNA ligase
Indicate whether each of the following statements about eukaryotic cells is true (T) or false (F). (10)
___ They have three distinct RNA polymerases.
___ Their mRNAs are generally synthesized by RNA polymerase I.
_____ RNA polymerase III synthesizes only rRNAs.__
______The 5S rRNA is synthesized by RNA polymerase I.
___ Their RNA polymerases initiate transcription at specific promoter sites on the DNA
T,F,F,F,T
Describe in words (not using structures) the important features of the structures present on the 5' and 3' ends of mature (processed) eukaryotic mRNAs. (8)
At the 5' end, there is a guanosine cap joined to the 5'-terminal nucleotide through a 5' to 5' triphosphate group, methylated on N-7.
At the 3' end is the poly(A) tail consisting of a run of 80–250 adenylate residues.
Briefly describe the role of the following components in bacterial protein synthesis. (12)
(a) Initiation factor 2 (IF-2)
(b) Shine-Dalgarno
(c) Peptidyl transferase
(d) Release factors
(e) Elongation factor G (EF-G)
(f) tRNAfMet
(a) IF-2 is a protein factor that, when bound to GTP, brings the fMet-tRNAfMet to the initiation complex.
(b) 16S RNA is a component of the small (30S) subunit. It contains a sequence complementary to the Shine-Dalgarno sequence in the mRNA, and helps to line up the mRNA initiation AUG codon on the ribosome.
(c) Peptidyl transferase is a ribozyme in the 50S ribosomal subunit. It catalyzes formation of each peptide bond as the ribosome moves along the mRNA.
(d) Release factors are proteins that bring about the release of the finished polypeptide when the ribosome encounters a termination codon in the mRNA.
(e) EF-G participates in the translocation of the ribosome down the mRNA by one codon after each peptide bond is formed.
(f) tRNAfMet is the transfer RNA that initiates protein synthesis by inserting the first amino acid (fMet) in every prokaryotic protein.
Briefly describe the five major groupings of amino acids (10).
Amino acids may be categorized by the chemistry of their R groups:
(1) nonpolar;
(2) polar, uncharged;
(3) aromatic
(4) positively charged;
(5) negatively charged.
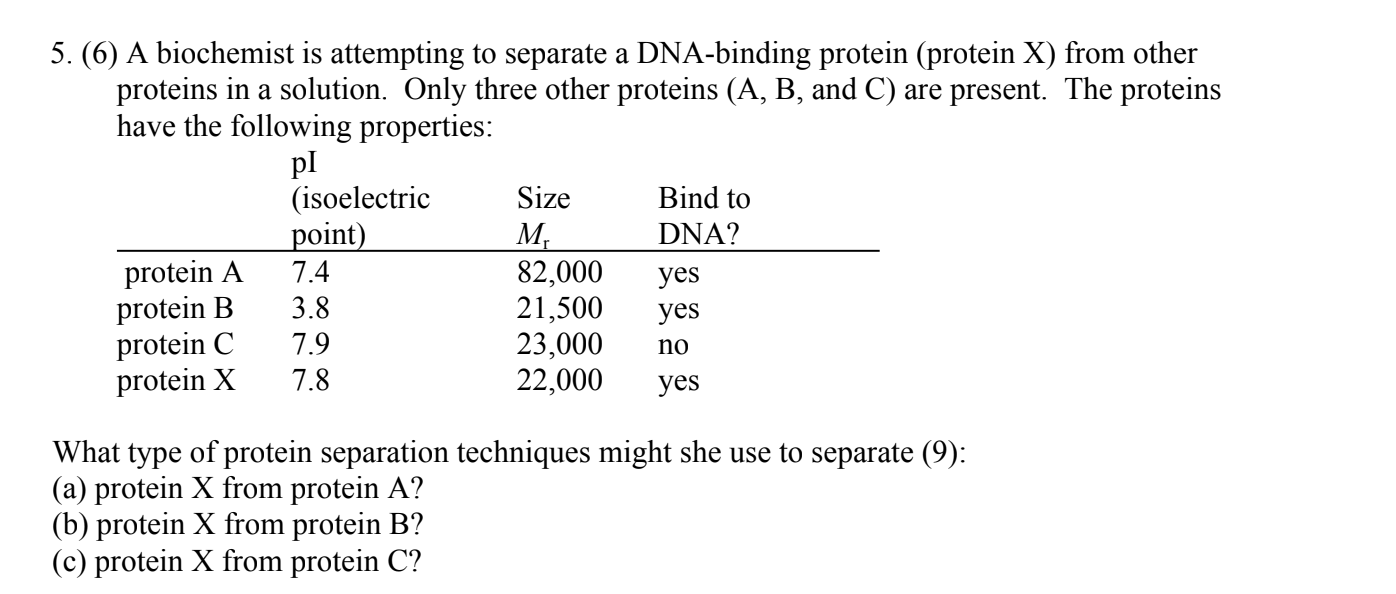
(a) Size-exclusion (gel filtration) chromatography to separate on the basis of size;
(b) ion-exchange chromatography or isoelectric focusing to separate on the basis of charge;
(c) specific affinity chromatography, using immobilized DNA.
Match the following list of RNAs with their functions.
A. mRNA 1. adaptor for protein synthesis
B. rRNA 2. codes for proteins
D. snRNA 3. components of ribosome
E. tRNA 4. splicing of RNA transcripts
2, 3, 4, 1
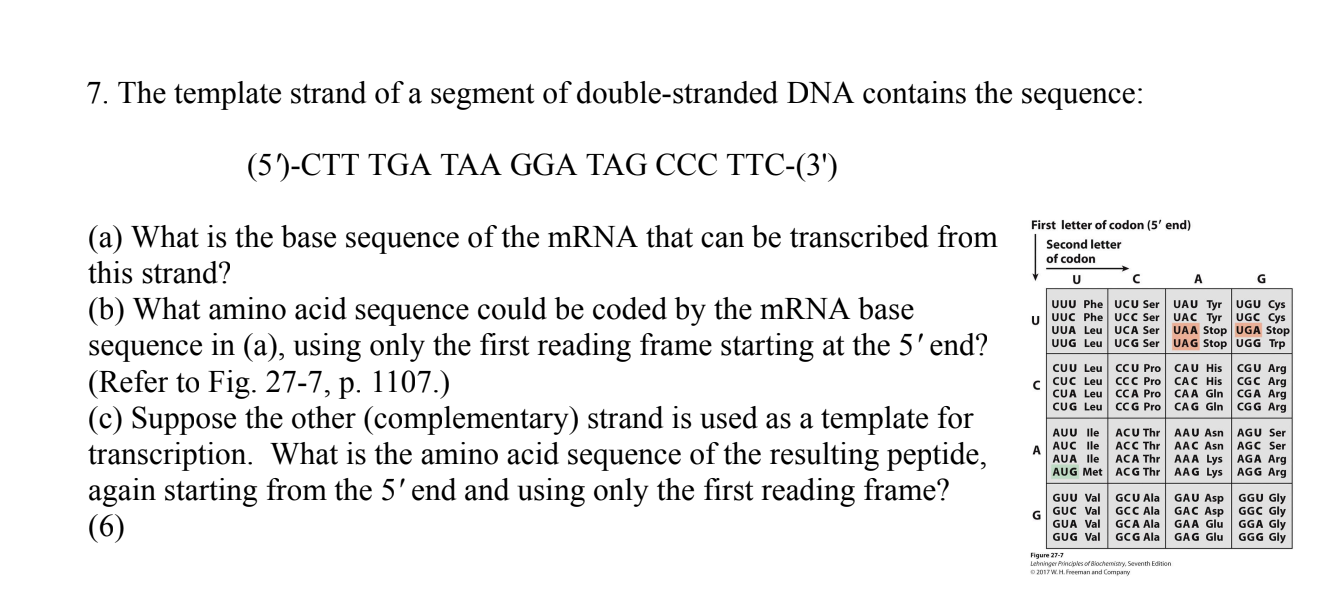
(a) (5')GAA GGG CUA UCC UUA UCA AAG(3')
(b) Glu-Gly-Leu-Ser-Leu-Ser-Lys
(c) The codons translate to Leu-Stop-Stop. No peptide would be produced because of the stop codons.
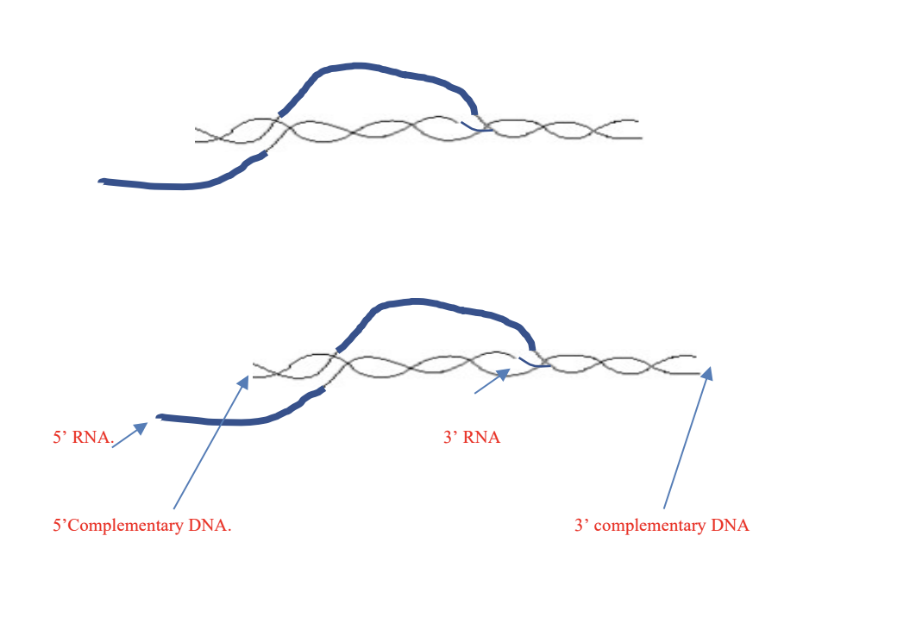
Below, an RNA molecule is being transcribed from a strand of DNA. Indicate the 5' and 3'
ends of the RNA molecule and of the strand of DNA that is complementary to the RNA
molecule. In which direction is synthesis occurring? (5)
Indicate whether each of the following statements is true (T) or false (F). (12)
Bacterial mRNA is broken down within a few minutes of its formation in E. coli.
Bacterial mRNA consists only of the bases that code for amino acids.
Bacterial mRNA normally occurs as a double-stranded structure, with one strand containing codons, the other containing anticodons.
Bacterial mRNA can be translated while it is still being synthesized.
T, F, F, T
Describe how you would determine the Ka (association constant) for a ligand and a protein.
Yo would perform an experiment with a fixed amount of protein and varying amounts/concentrations of ligand until an equilibrium is reached. Then, plot a graph with ligand concentration vs fraction of proteins with a ligand bound to it. The ligand concentration is 1/Ka when the fraction is at 0.5.
Michaelis-Menton Kinetics is sometimes referred to as “saturation” kinetics. Why?
When [S] becomes very high, an enzyme molecules active site will become occupied with a new substrate molecule soon as it releases a product. Therefore, at very high [S], V0 does not increase with additional substrate, and the enzyme is said to be “Saturated” with substrate
An enzyme catalyzes a reaction at a velocity of 20mol/min when the concentration of substrate (S) is 0.01 M. The Km for this substrate is 1×10^-5M. Assuming that Michaelis Menten kinetics are followed, what will the reaction velocity be when the concentration of S is 1×10^-5M?
Vo = 20 x (1×10^-5)/(1×10^-5)(1×10^-5) = 10 mmol/min
Enzymes with a cat/K ratio of about 10^8 M^-1s^-1 are considered to show optimal catalytic efficiency. Fumarase, which catalyzes the reversible-dehydration reaction. Fumarate + H20 —> malate has a relation of turnover number to Michaelis-menten constant, (cat/Km) of 1.6×10^8 for the substrate fumarate and 3×10^7 for the substrate malate. because the turnover number for both substrates is nearly identical, what factors might be involved that explain the different ratio for the two substrates?m
Since both substrates are nearly identical, a difference in Km may explain the different ratio. The Different Kms may be due to the strength of the enzyme and the binding affinity.
(a) what is the effect of pH on the binding of oxygen to hemoglobin (the Bohr Effect)? (b) Briefly describe the mechanism of this effect.
(a) The affinity decreases with decreasing pH. (b) At lower pH (i.e., higher H+ concentration), there is increasing protonation of protein residues such as histidine, which stabilizes the low affinity conformation of the protein subunits.
Name four factors (bonds or other forces) that contribute to stabilizing the native structure of a protein and describe one condition or reagent that interferes with each type of stabilizing force.
Disulfide bonds - mercaptoethanol
Hydrogen bonds - pH extremes
Hydrophobic interactions - Detergent & urea
ionic interactions - changes in pH
Why are glycine and proline often found within a beta turn?
Glycine is common in beta turns because it does not have a beta carbon and as a result no side chain. This gives it more flexibility when it comes to the angles that it can occupy in loops. Proline is found most often in a beta turn because its side chain attaches to the protein backbone two times instead of once, which makes it very stable to introduce into loops but not into anywhere else.
A biochemist obtains the following set of data for an enzyme that is known to follow
Michaelis-Menten kinetics.
Substrate Initial
concentration velocity
(µM) (µmol/min)
1 49
2 96
8 349
50 621
100 676
1,000 698
5,000 699
(a) Vmax for the enzyme is 700. Explain in one sentence how you determined Vmax.
I determined the Vmax for the enzyme by determining the value that the initial velocity
approaches but never meets, which in this case would be 700 (µmol/min).
(b) Km for the enzyme is 8. Explain in one sentence how you determined Km.
Km for the enzyme can be determined by finding the concentration at which half of the Vmax was attained, which would be 8 um.