M5 Poole- Prokaryotic gene regulation
1/11
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
12 Terms
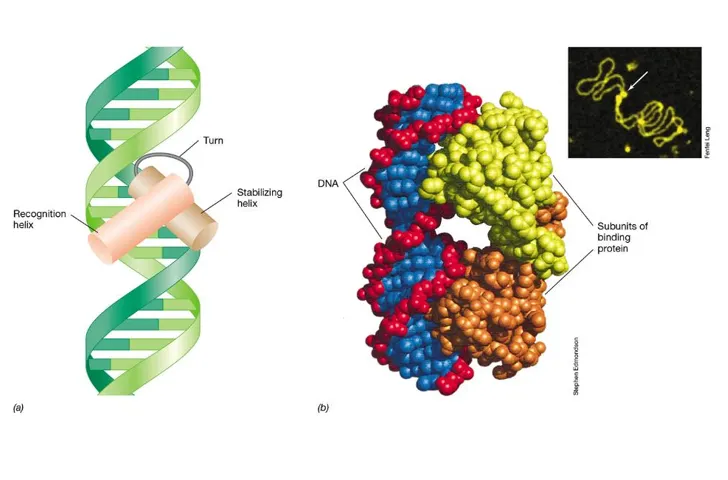
what is the most common structure of gene regulators?
regulators are usually dimers (two very similar protein monomers)
the helix-turn-helix motif is a very common motif
these form two domains: a stabilising helix and a recognition helix
what are the three types of gene regulation?
negative regulation always has a repressor which binds to the operatore (downstream of the promoter) and stops RNA polymerase:
negative regulation resulting in repression eg. lac operon
a corepressor binds to the repressor protein and causes it to attach to the operator so that RNA polymerase can’t transcribe DNA
negative regulation resulting in induction
an inducer binds to the repressor and causes it to detach from the operator, so that RNA polymerase can transcribe DNA
positive regulation always has an activator which binds to the activator binding site (upstream of the promoter) and permits RNA polymerase:
positive regulation resulting in activation
an inducer binds to the activator protein which attaches to the activator binding site and allows RNA polymerase to attach to the promoter so that it can transcribe DNA
the binding site can be quite far upstream of the promoter, so the dna needs to bend

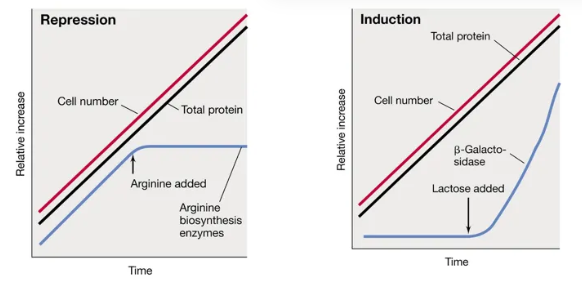
what is the difference between the two types of negative gene expression and when are they used most? (with graphs)
anabolic genes (biosynthetic) are typically subject to repression by the end-product
the repressor protein is only attached when the end-product (the corepressor) is present
when arginine is present, the repressor protein attaches and arginine biosynthesis enzymes stop working
catabolic genes (degradative) are typically induced by a substrate
the repressor protein is only detached when the substrate (the inducer) is present
when lactose is present, the repressor protein detaches and the lactase enzymes start working
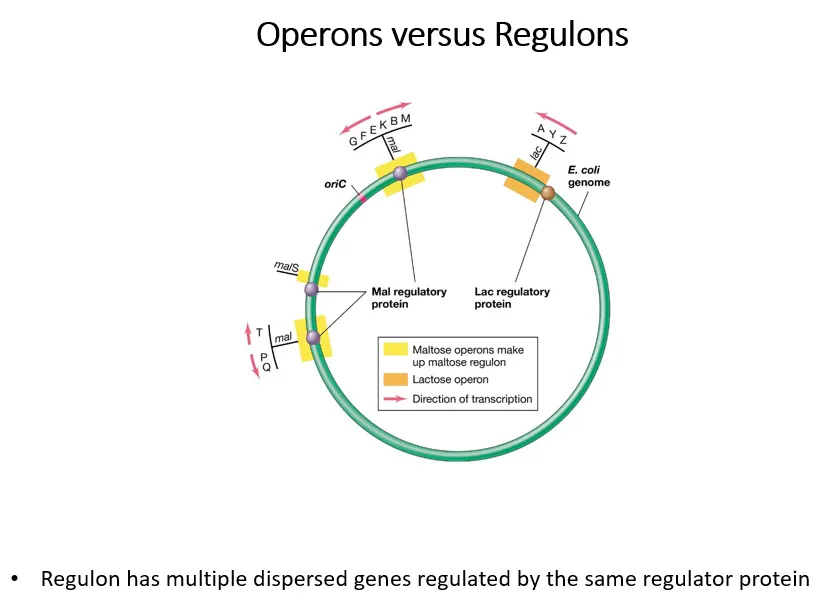
what are regulons and operons?
operons are a group of genes controlled by a regulator protein
regulons are multiple groups of operons controlled by the same regulator protein
the yellow operons are part of the maltose regulon (all controlled by the mal regulatory protein), the orange operon isn’t part of a wider lac regulon
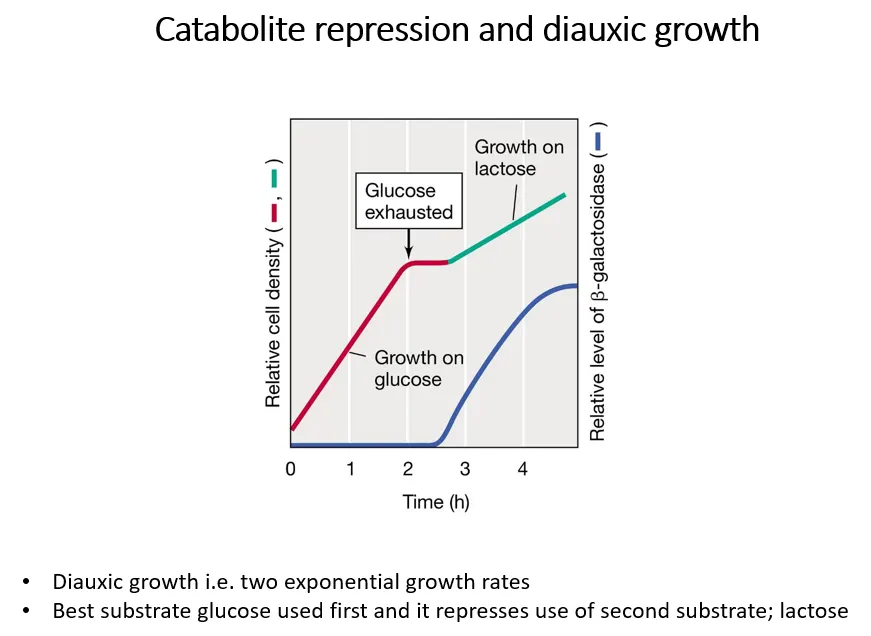
what is diauxic growth?
diauxic growth is shown when two metabolic substrates are present, eg. both glucose and lactose
glucose is a better substrate, so bacteria grow on it preferentially at first, then switch to lactose when it gets used up
if glucose is present, the lactase enzymes aren’t transcribed
when glucose runs out, bacterial growth stops while the lac operon is being induced to produce the lactase enzymes
this means the lac operon is under two levels of regulation:
positive regulation dependent on glucose concentrations
negative regulation dependent on lactose concentrations
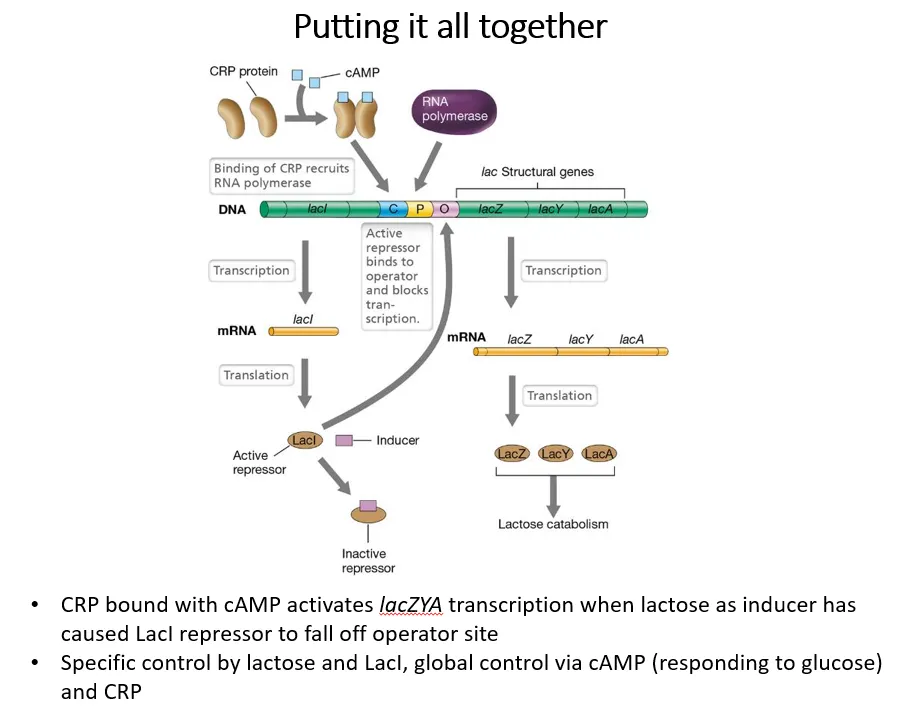
how do glucose and lactose concentrations regulate the lac operon?
positive regulation dependent on glucose:
adenylate cyclase converts ATP into cyclic AMP
cAMP acts as an inducer, binding to CRP (cAMP receptor protein), which is an activator for the lac operon
glucose inhibits adenylate cyclase, so when glucose is present, little cAMP is produced
this means less CRP can bind to the activator binding site, so RNA polymerase can’t bind
negative regulation dependent on lactose:
lactose acts as an inducer for the LacI repressor protein
when lactose is present, the repressor detaches from the operator region, so RNA polymerase can bind
this means transcription is only possible if CRP is present, due to the absence of glucose, and if the LacI repressor is inactivated, due to the presence of lactose
how do bacteria regulate gene expression based on the environmental signals?
bacteria have many sensors, autokinases that tend to be membrane proteins
these can recognise different environmental signals and phosphorylate themselves using ATP
they can then transfer this phosphate onto a response regulator, which is typically a transcription factor
the phosphate acts as an activator or inducer on different genes
this is called a two-component system

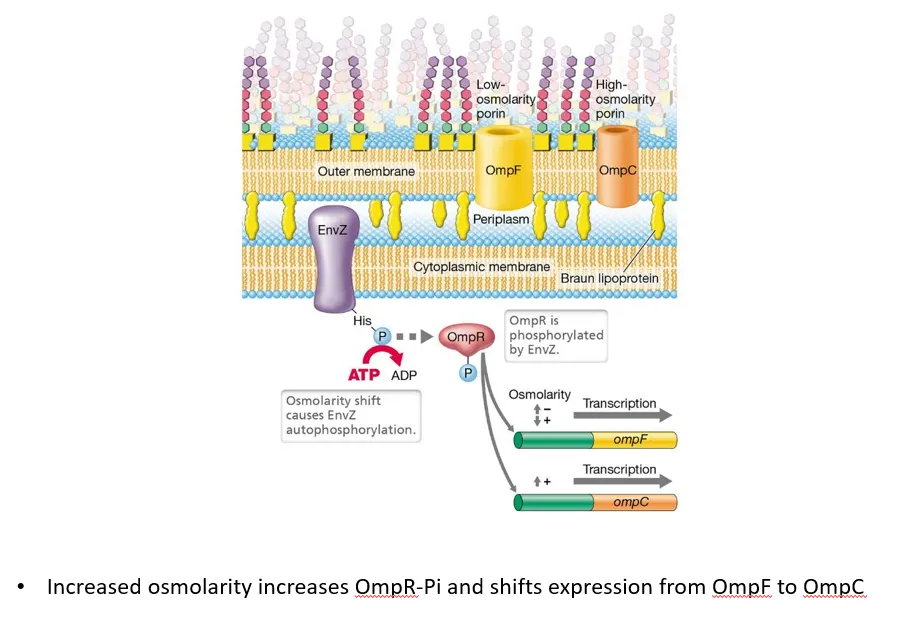
what is an example of a two-component system in G-ve bacteria?
the sensor envZ, a transmembrane protein in the cytoplasmic membrane of G-ve bacteria, detects changes in the osmotic potential of the periplasm
when the osmolarity shifts, it autophosphorylates, then transfers the phosphate to the ompR regulator protein
low osmolarity (high solute concentration outside the cell) causes ompR to induce ompF (large protein channel) production and repress ompC (small protein channel), so more solutes diffuse in
high osmolarity (low solute concentration outside the cell) causes ompR to repress ompF production and induce ompC, so more water diffuses in
this helps achieve the correct osmotic pressure inside the cell, to maintain turgor
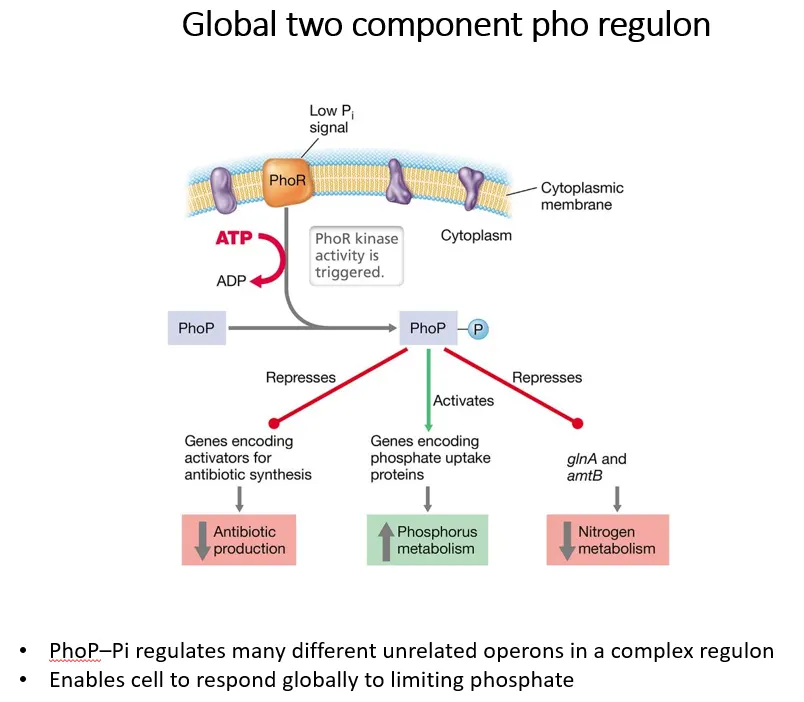
what is an example of a two-component regulon in G+ve bacteria?
the sensor PhoR in the cytoplasmic membrane in G+ve bacteria detects low phosphate in the environment and autophosphorylates using ATP
it then ???????
what is an example of transcriptional control in archaea?
amino acids are normally produced by the transamination of oxaloacetate by glutamate, also producing alpha-ketoglutarate
if nitrogen is limiting, glutamate isn’t reformed from a-ketoglutarate
so if a-ketoglutarate is high, nitrogen-acquiring genes must be turned on
a-ketoglutarate is an inducer, which binds to the nrpR repressor protein and causes it to drop off the TATA and BRE boxes so TBP and TFB activators can bind, allowing RNA polymerase to attach and transcribe the DNA

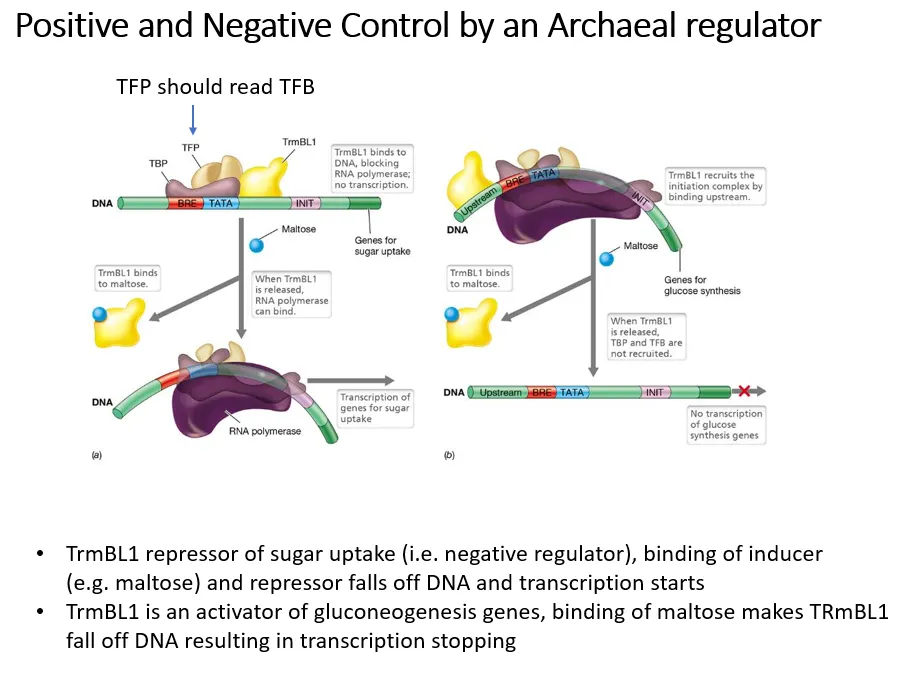
what is an example of a regulator that can both positively and negatively control genes in archaea?
negative control by induction:
trmBL1 acts as a repressor of sugar uptake genes
when sugars are present, maltose acts as an inducer and binds to trmBL1 so that it drops off
this means transport systems to take up the sugars can be transcribed
positive control:
trmBL1 also acts as an activator of gluconeogenesis genes
when sugars are present, maltose binds to trmBL1 and causes it to drop off
this means the gluconeogenesis genes can’t be transcribed when there are sugars already present
explain what sigma factors are
sigma factors are cofactors for RNA polymerase- they recognise and bind to the promoter sequence, then recruit RNA polymerase to begin transcription (and form a holoenzyme together), then drop off
the main sigma factor is sigma 70, which is produced from the rpoD gene (the housekeeping gene)
this binds to pribnow boxes- the more similar the sequence is to the TATA box, the more often sigma 70 will bind and the more the gene will be expressed