BIS 002A MOCK-MT3-WINTER 2025
1/24
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
25 Terms
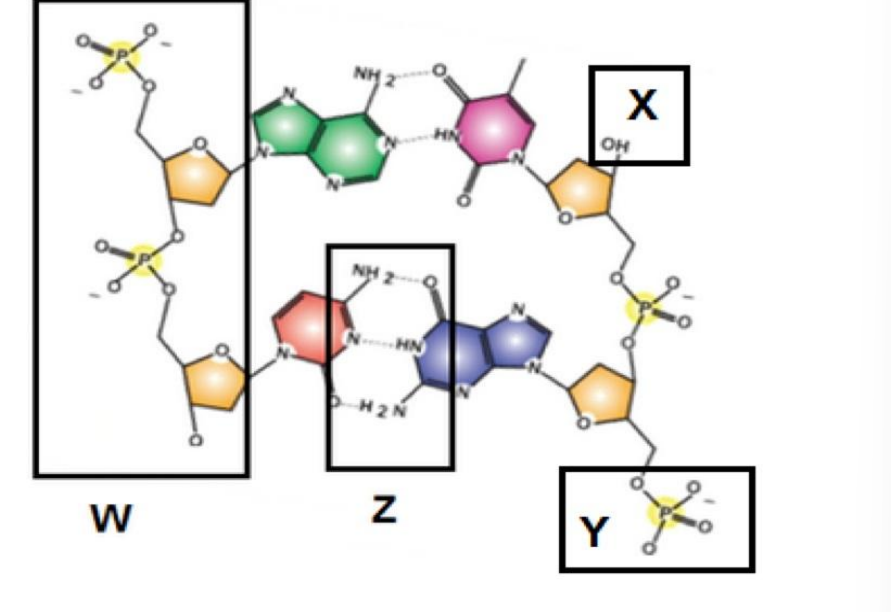
The backbone of this polymer is shown in. The functional group denoted with is necessary for the addition of the next monomer
W, X
Explanation:
The backbone of DNA is made of sugar-phosphate linkages.
The functional group necessary for polymerization is the 3’-OH (hydroxyl) group of the sugar, which allows the next nucleotide to be added via phosphodiester bond formation.
The addition of a nucleotide to a growing DNA strand requires energy. One of the products of this reaction is a DNA polymer with additional nucleotide. The other product is
PPi
Explanation:
DNA polymerization involves the hydrolysis of deoxynucleotide triphosphates (dNTPs).
When a nucleotide is incorporated, pyrophosphate (PPi) is released, which provides energy to drive the reaction forward.
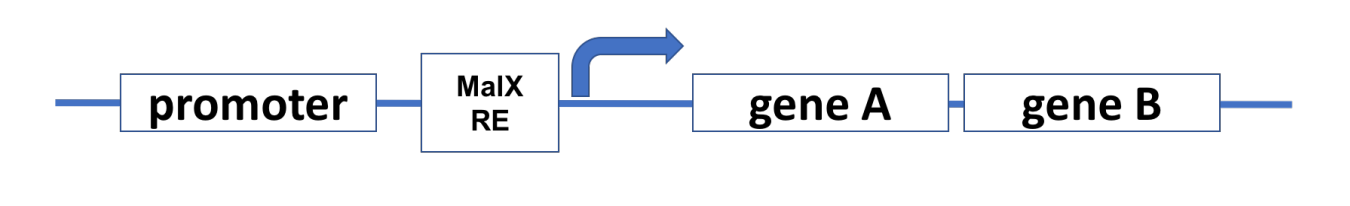
The E. coli chromosome contains the malanose operon (shown below). The malanose operon has a “strong promoter,” which is negatively regulated by MalX transcription factor. MalX protein exerts its influence by binding to a regulatory element called the MalX RE.
An E. coli strain is isolated that has a mutation prevents the production of the MalX protein. What effect will this mutation have on transcription of the malanose operon genes A and B
Transcription of the malanose operon will occur
the cell will not be able to turn the malanose operon OFF using MaIX
Explanation:
MalX is a repressor that inhibits transcription of the malanose operon.
If MalX is missing, the operon cannot be turned OFF, meaning transcription will always occur.
Since MalX is necessary for repression, the cell loses the ability to shut down transcription.
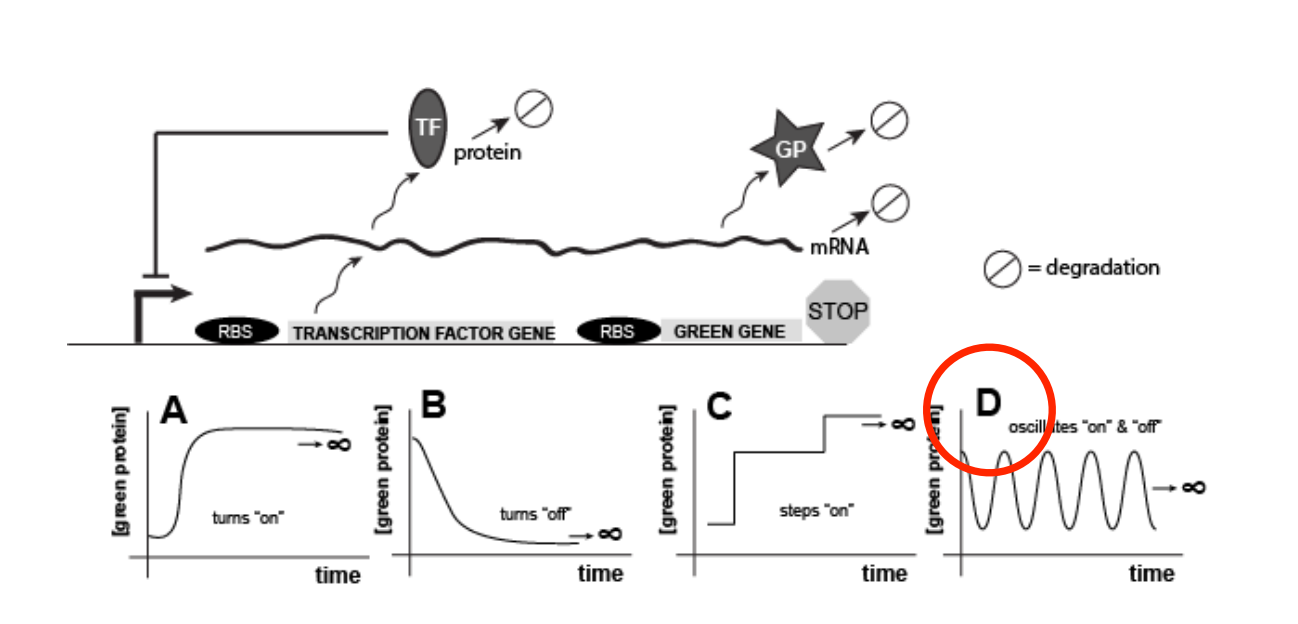
Given the picture of the genetic circuit below, which of the following graphs of green protein expression versus time would be the most reasonable behavior to expect from cells expressing this genetic element? Assume that ∞ indicates that the observation extends over infinitely long periods of time.
Explanation:
DNA replication is semi-conservative, meaning each new DNA molecule consists of one old (template) strand and one newly synthesized strand.
Both the leading and lagging strands serve as templates for the synthesis of new complementary strands.
DNA polymerase reads the template strand in the 3' → 5' direction while synthesizing the new strand in the 5' → 3' direction.
If a double helical strand of DNA is analyzed and contains 35 percent T bases, what percent of the strand contains C bases
15
EXPLANATION:
Chargaff’s rule states A = T and G = C.
If T = 35%, then A = 35%.
This leaves 30% for G + C.
Since G and C are equal, each must be 15%.
The association of DNA with histones typically affects transcription by:
. decreasing transcription of a few genes, while not influencing most others.
Explanation:
Histones wrap DNA into chromatin, making it less accessible for transcription.
This reduces transcription by physically blocking RNA polymerase.
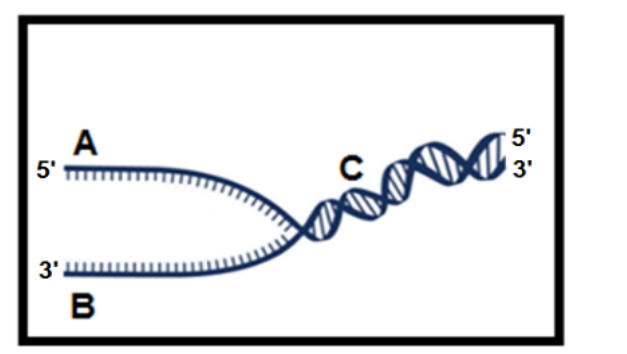
Use the replication fork figure to answer this question. The replication fork is moving from left to right. The strand labeled________ is the template for the _______ and is created from __________. Okazaki fragments will be located on strand _____.
B, leading strand, 5’ to 3’, A
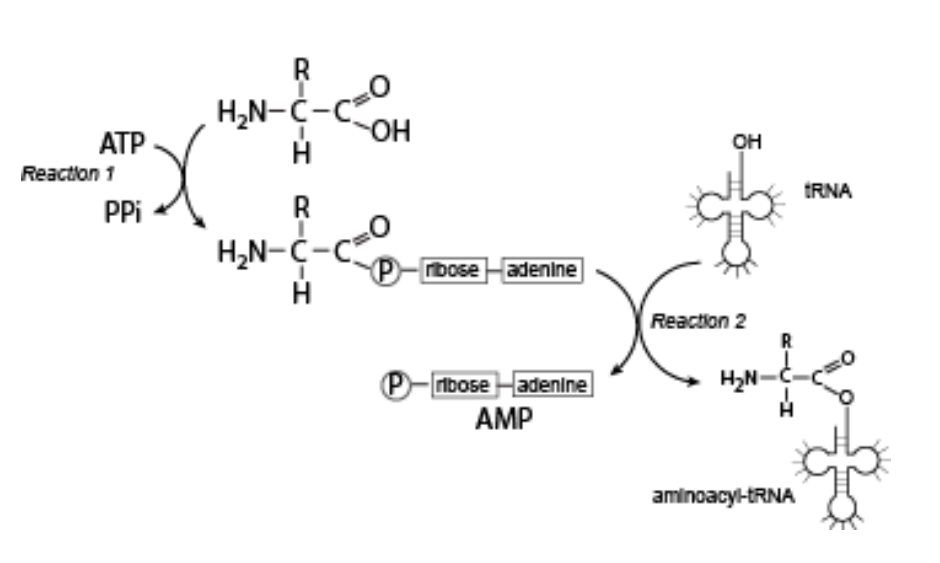
The reactions shown in the figure below are carried out by the enzyme aminoacyl-tRNA synthetase. In this scheme, the formation of a covalent bond between AMP and an amino acid serves to:
Temporarily “store” some of the energy released by ATP hydrolysis
Explanation:
The reaction forms an aminoacyl-AMP intermediate, which stores energy.
This energy is later used to attach the amino acid to the tRNA, making it ready for translation.
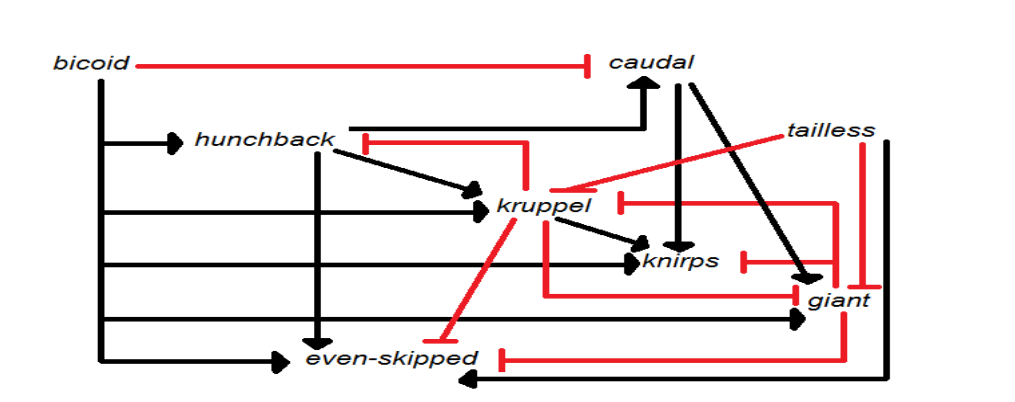
Using the gene diagram below, which statement is true
Kruppel is directly regulated by 4 different proteins
EXPLANATION:
Red lines typically represent exons (coding sequences).
Black lines typically represent introns (noncoding sequences).
The open reading frame (ORF) starts within the first red exon and continues across subsequent red regions.
Eukaryotic cells divide by mitosis and meiosis. Below are a set of five characteristics that describe one or both of these modes of cell division. Consider this list and answer the question below using these five possibilities. Which TWO of the criteria apply uniquely to MITOSIS?
I. Can occur in either haploid or diploid cells.
II. Attempts to assure that daughter cells are genetically identical to the parent cells.
III. Involves pairing between homologous chromosomes.
IV. Generates progeny with a variety of genotypes.
V. Produces progeny cells with half the amount of DNA as the parent.
I and II
Explanation:
Mitosis occurs in both haploid and diploid cells.
The goal of mitosis is to produce genetically identical daughter cells.
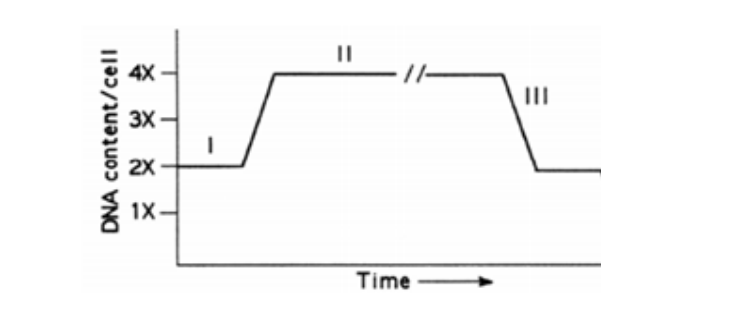
The figure below represents changes in the DNA content of a cell as it progresses through the cell cycle.
The above figure represents a population of _____________ cells going through ____________ to generate _______________
diploid; mitosis; diploids
Explanation:
Meiosis starts with diploid cells.
It reduces chromosome number by half, producing haploid gametes.
Within the human population, 200 different alleles exist for a specific locus. Each person carries at most ______ alleles for this locus
2
Explanation:
Humans are diploid, meaning each individual inherits one allele from each parent.
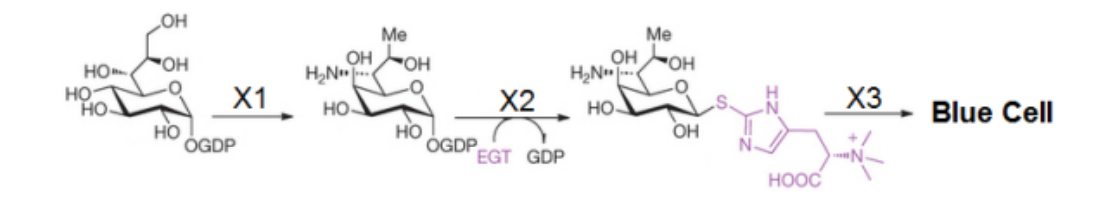
The Entoloma hochstetteri mushroom from New Zealand produces a beautiful blue pigment as an end product to a metabolic pathway shown below.
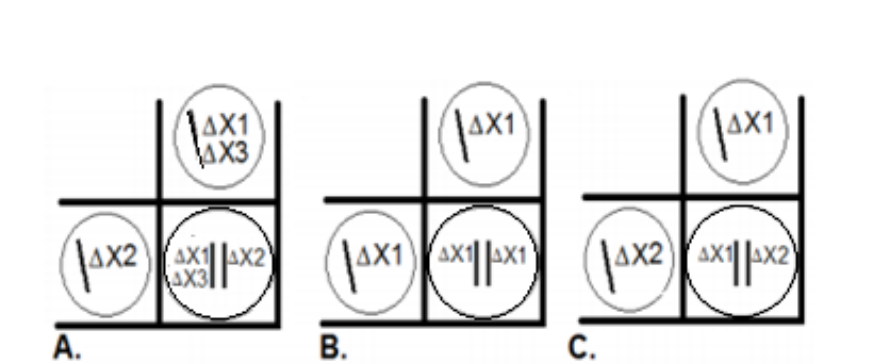
You currently have two haploid Entoloma hochstetteri, both are white. You mate them to form diploid offspring. You find that the diploid offspring is blue! The images below are Punnett squares showing the chromosomes of the haploid parents and the chromosomes of the resulting diploid. Which image below accurately portrays a cross that could account for the diploid blue cell
A and C
EXPLANATION:
Why Images A and C Are Correct:
These images likely show one parent missing one functional gene, while the other parent has a functional copy of that gene.
This means that the diploid offspring inherits at least one functional allele for every gene in the pathway, enabling blue pigment production.
Key Concept for Exams:
"Complementation restores function in diploids."
"Haploid mutants alone = no pigment, diploid = pigment restored."
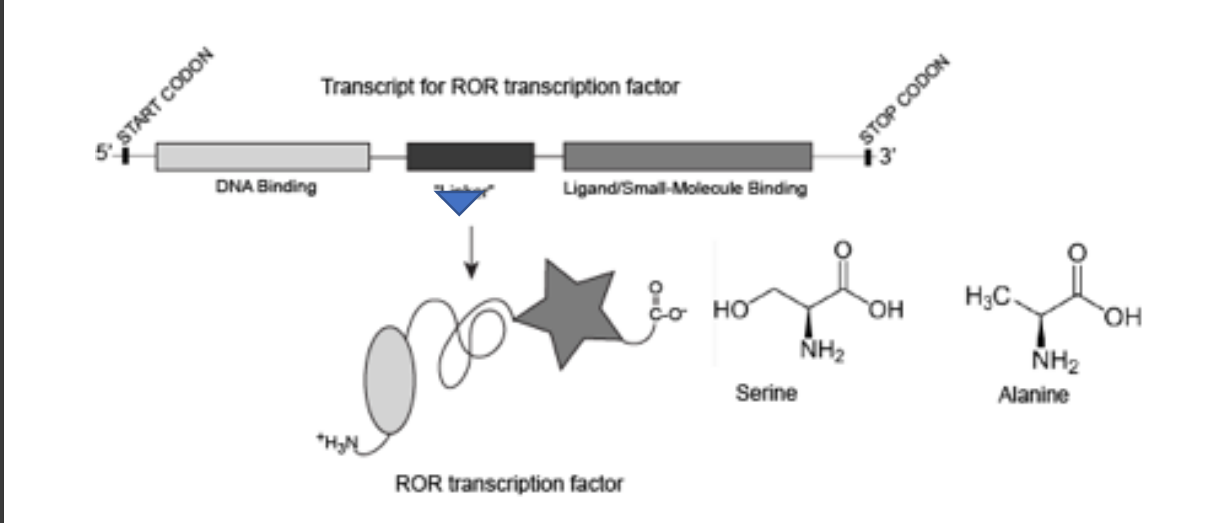
14) The protein ROR, shown below, is a transcription factor that binds small molecules. The binding of small molecules acts like a switch that leads to a change in tertiary structure that helps recruit other proteins to the promoter, thereby up-regulating transcription of target genes.
A single mutation (ARROW in the figure) changes a codon encoding a serine in the “linker” region of ROR into a codon encoding an alanine. The mutant ROR protein no longer up-regulates transcription in the presence of ligand. The simplest plausible/ reasonable explanation for this is
The serine was involved in a hydrogen bond with another part of the protein that was important for changing the shape of the DNA binding domain.
Explanation:
Serine can form hydrogen bonds, but alanine lacks a hydroxyl group.
This likely disrupts the protein’s ability to change conformation, preventing DNA binding.
The telomerase enzyme consists of an RNA molecule and a protein. What is the role of the RNA molecule in telomerase?
Acts as a template
Has a complementary sequence to the telomere repeat
Explanation:
Telomerase uses its RNA component to extend the ends of chromosomes by acting as a template for new DNA synthesis.
The structure of linear s present a special problem/challenge for DNA replication because . . . .
The 3’ “end” of the lagging strand offers no spot for the enzyme primase to prime for synthesis of the last bit of chromosome
Explanation:
DNA polymerase requires a primer to start synthesis.
The end-replication problem means that the lagging strand cannot be fully replicated.

Which of the following sites would you predict to be present in the gene encoding a small regulatory RNA molecule and what would be their order?
1,3,4
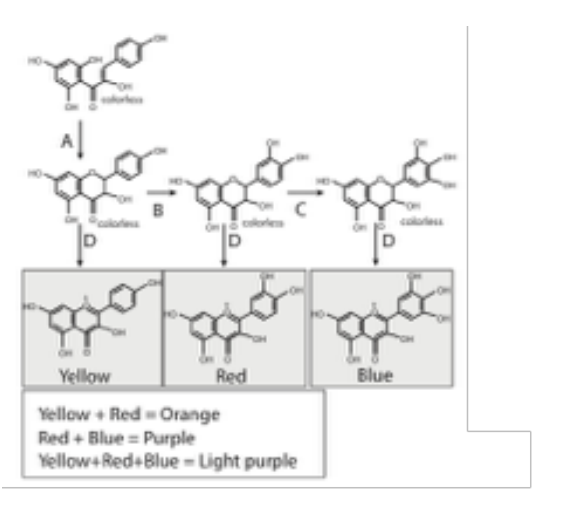
The figure to the right diagrams a metabolic pathway that might controls the color of flowers in certain plants: The genotype of a diploid wild-type flower is AA BB CC DD, and its phenotype is light purple. Alleles A, B, C, D each encoded enzymes that catalyze the eponymous reactions in the Figure. a, b, c, and d are loss-of-function alleles. What will the mostly likely phenotype of the AA Bb cc DD double mutant?
ORANGE
An mRNA has the sequence 5'-AUGAAAUCCUAG-3'. What is the template DNA strand for this sequence?
5'-CTAGGATTTCAT-3'
Explanation:
mRNA is complementary and antiparallel to the template strand.
The correct answer must have complementary base-pairing in reverse order.
Gene Q is found in a eukaryotic cell and has a single 1kb intron in its middle. A DNA deletion removes 10 base pairs from the middle of this intron. The most likely consequences of this mutation on the ORF encoded by the mature RNA of this gene are:
none, intron is not part of the ORF
Explanation:
Introns are removed before translation.
A small intron deletion does not affect the protein sequence.

Two different point mutations were discovered in the promoter sequences in two mutant bacteria (mutant 1 and mutant 2). Their activities were measured and compared to the wild-type (non-mutant) promoter (see table below). Which of the following statements is/are true?
A. The number of proteins produced per mRNA depends upon the strength of the promoter.
B. Changing the sequence of the promoter has little to no effect because both mutants can still produce mRNAs and proteins.
C. Changing the 3rd nucleotide in the promoter does not lower the strength of the promoter as much as changing the 6th nucleotide in the promoter sequence.
Changing the 3rd nucleotide in the promoter does not lower the strength of the promoter as much as changing the 6th nucleotide in the promoter sequence
EXPLANATION:
Codon Redundancy (Degeneracy of the Genetic Code)
The genetic code is redundant, meaning multiple codons can specify the same amino acid.
Example: Leucine (Leu) is encoded by CUU, CUC, CUA, and CUG.
This redundancy helps minimize the impact of mutations, as changes in the third base often do not alter the amino acid (known as the wobble effect).
In bacteria, it is possible to encode a protein with the same amino acid sequence by using many different DNA sequences. What is the best/most common explanation?
The redundancy of the genetic code enables the use of multiple codons for the same amino acid
Explanation:
The genetic code is degenerate, meaning multiple codons can specify the same amino acid.
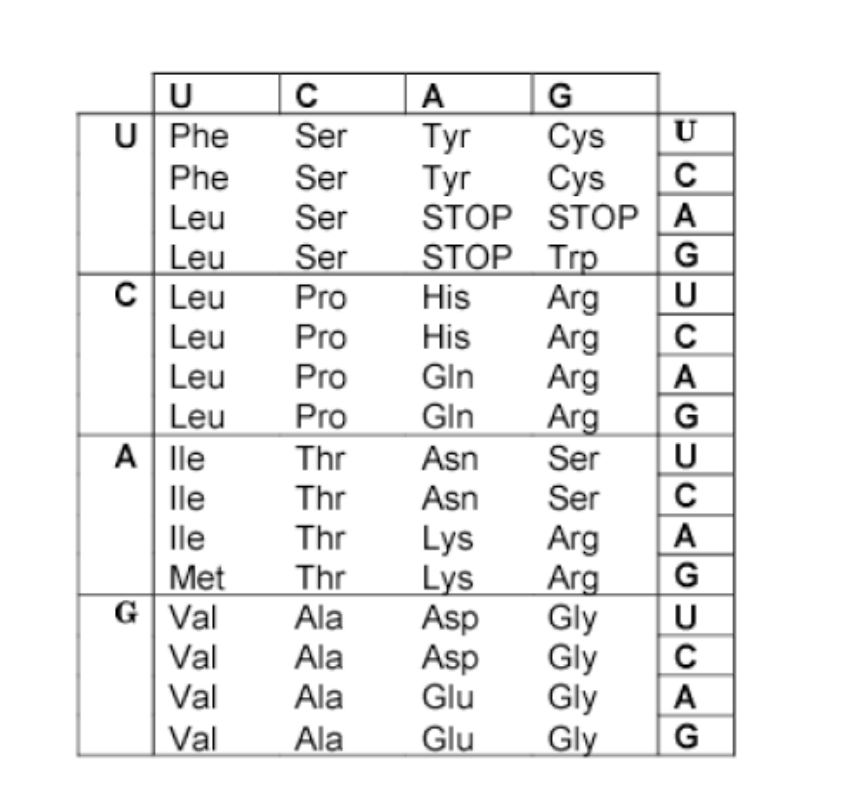
)Based on the DNA strand below, and assuming the promoter for this gene is located to the left, which protein sequence below does this sequence code for? (Assume the first nucleotide above is the start of the first codon.)
5’-ATGTTGAAAATGCCGTAGAGGC-3’
Met-Leu-Lys-Met-Pro
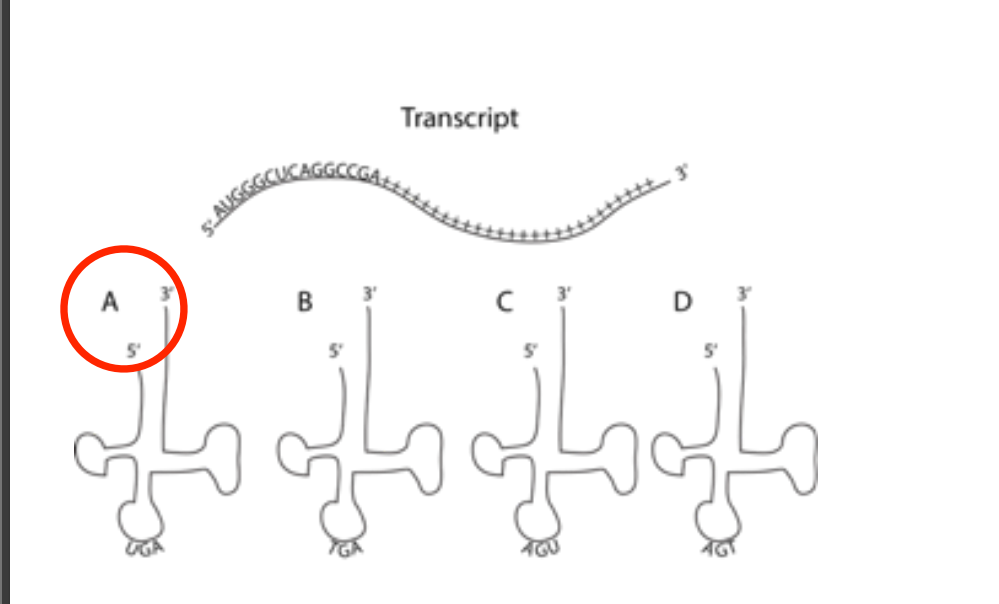
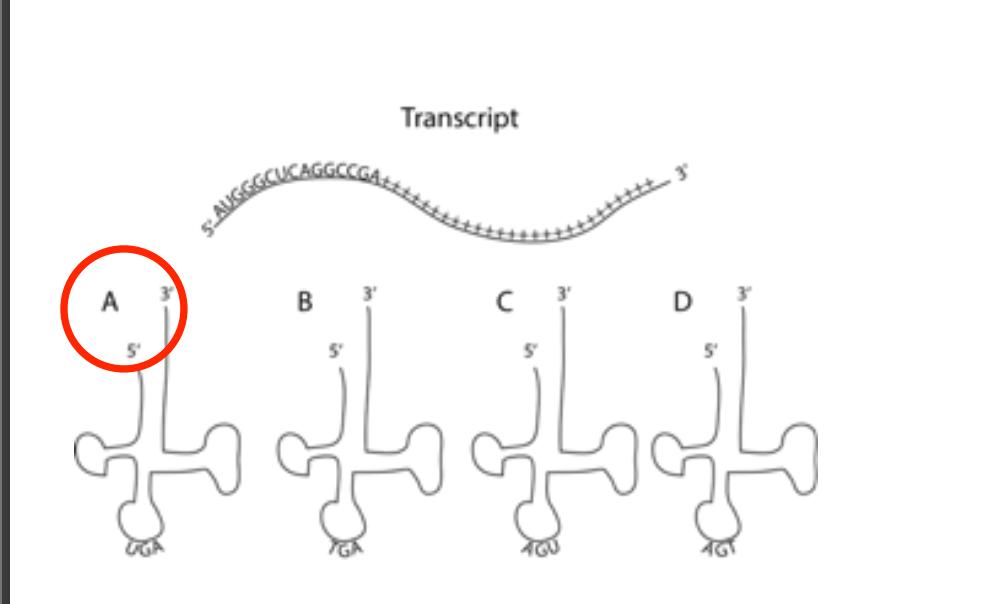
The figure below depicts a transcript. The (+) sign just indicates any random nucleotide. Which of the tRNA molecules depicted below it are the best match for the third codon?
FIGURE A
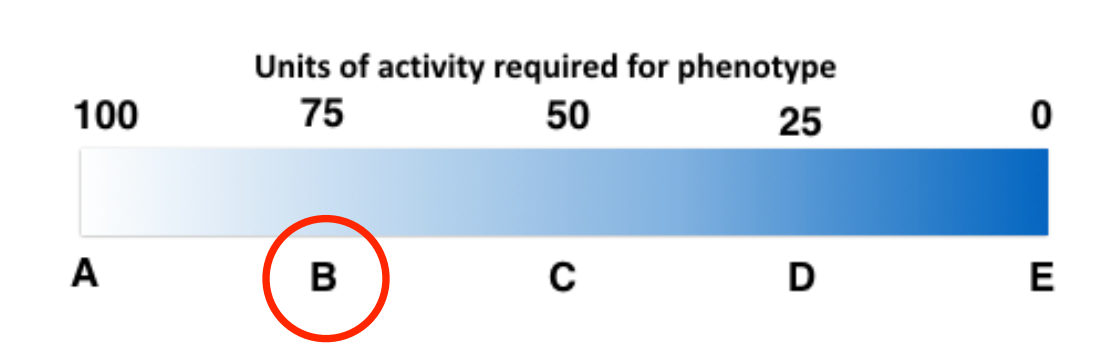
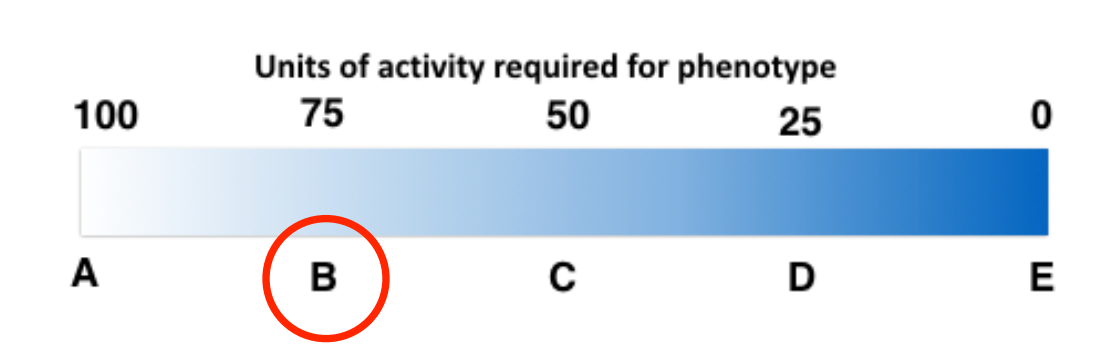
25) Imagine the following partially hypothetical scenario: A typical sized yeast cell (haploid or diploid) requires a minimum of 100 units of enzyme activity from the protein encoded by the ade2 gene in order to keep a white phenotype when the purine biosynthetic pathway is operating .
• The wild-type ade2 allele produces 100 units of activity.
• A haploid yeast expresses allele ade2-a that encodes an enzyme that provides 50 units of activity.
• A second haploid yeast expresses allele ade2-ts that also produces 50 units of activity but loses ½ of its function when yeast are grown at low temperature.
The expected phenotype (given by the letter below) of the diploid yeast grown at low temperature (assuming equal expression of each gene) is: