BI216 exam 3
5.0(1)
5.0(1)
Card Sorting
1/56
Earn XP
Description and Tags
Study Analytics
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
57 Terms
1
New cards
DNA structure
GC (triple bonds) stronger than AT (double H bonds)
antiparallel strands
major and minor groove
one turn of helix - 10bp
antiparallel strands
major and minor groove
one turn of helix - 10bp
2
New cards
stem loop
pairing of ssDNA/RNA
4 bases needed for loop
4 bases needed for loop
3
New cards
Recombination
ds-breaks lead to recombination
meiosis - promotes genetic diversity (by Spoll protein)
mainly in DNA repair (Ds-breaks lethal if not repaired)hol
meiosis - promotes genetic diversity (by Spoll protein)
mainly in DNA repair (Ds-breaks lethal if not repaired)hol
4
New cards
holiday juction resolution
via RuvC
5
New cards
branch migration
via RuvAB
6
New cards
gene conversion (definition)
number of genotypic outcomes is not equal as in starting genotype
7
New cards
gene conversion via mismatch repair/recombination
mismatch repair cant distinguish a correct strand → after recombination repair strand is equally as probable
each mismatch is resolved independently
each mismatch is resolved independently
8
New cards
mutation (definition)
heritable change in DNA sequence (DNA damage, mismatch/misrepair are not mutations)
9
New cards
Transition
no change in total number of base pairs
purine/pyrimidine → purine/pyrimidine (GC→AT)
purine/pyrimidine → purine/pyrimidine (GC→AT)
10
New cards
Transversion
purine/pyrimidine → pyrimidine/purine (AT → TA, AT → GC)
11
New cards
missense mutation
wrong amino acid produced bc of base substitution
12
New cards
nonsense mutation
stop codon produced instead of amino acid
13
New cards
Indels
change in number of base pairs
insertion or deletion
in coding regions → frame shift mutation (change in reading frame during translation)
insertion or deletion
in coding regions → frame shift mutation (change in reading frame during translation)
14
New cards
isolating mutants
selection - only mutants grow
screening - phenotypical differences between wild and mutant
screening - phenotypical differences between wild and mutant
15
New cards
mutagenesis
if repair doesnt occur
two rounds of replication for mutagenesis to occur
two rounds of replication for mutagenesis to occur
16
New cards
17
New cards
base excision repair
removes small lesiosns
1. DNA glycosylase cleaes altered base from sugar nucleotide → AP site
2. AP endonuclease makes a nick in backbone at AP site
3. DNA polymerase fills in gap by copying undamaged strand
4. DNA ligase seals nick in the backbone
1. DNA glycosylase cleaes altered base from sugar nucleotide → AP site
2. AP endonuclease makes a nick in backbone at AP site
3. DNA polymerase fills in gap by copying undamaged strand
4. DNA ligase seals nick in the backbone
18
New cards
nucleotide excision repair
removes large lesions
1. exposure to UV light leads to damage
2. thymine dimer forms
3. UvrB and C endonucleases nick strand containing dimer
4. damaged fragment is released from DNA
5. DNA polymerase fills in gap with new DNA
6. DNA ligase seals repaired strand
1. exposure to UV light leads to damage
2. thymine dimer forms
3. UvrB and C endonucleases nick strand containing dimer
4. damaged fragment is released from DNA
5. DNA polymerase fills in gap with new DNA
6. DNA ligase seals repaired strand
19
New cards
non homologous end joining
repairs DNA damage/ds-breaks formed during G1
1. catalytic subunit (KO70, KO80, DNA-PKCS)
2. nuclease cleans up damaged ends
3. DNA ligase rejoins ds-ends
frequent occurance of mutation (deletions or insertions)
1. catalytic subunit (KO70, KO80, DNA-PKCS)
2. nuclease cleans up damaged ends
3. DNA ligase rejoins ds-ends
frequent occurance of mutation (deletions or insertions)
20
New cards
direction of RNA synthesis
5 → 3
21
New cards
direction of DNA copying
3 → 5
22
New cards
sigma subunit
recognizes -35 sequence and -10 sequence
separates DNA strands from -12 to +2
sigma must rearrange for RNAP to leave promoter and continue elongation
separates DNA strands from -12 to +2
sigma must rearrange for RNAP to leave promoter and continue elongation
23
New cards
termination of transcription (bacteria)
inversted repeat sequence
stem loop + multiple Us is a transcription termination signal
stem loop signals release of:
* DNA from RNA
* DNA from RNAP
* RNA from RNAP
need 4 bases for a loop
\
stem loop + multiple Us is a transcription termination signal
stem loop signals release of:
* DNA from RNA
* DNA from RNAP
* RNA from RNAP
need 4 bases for a loop
\
24
New cards
translation
mRNA to protein
tRNA mediate translation (each tRNA carries one particular amino acid)
tRNA mediate translation (each tRNA carries one particular amino acid)
25
New cards
tRNA
primary structure - tRNA sequence
secondary structure - clover leaf
secondary structure - clover leaf
26
New cards
start codon
AUG
27
New cards
stop codons
* UAA
* UAG
* UGA
* UAG
* UGA
28
New cards
open reading frame
open reading frame - no stop codon
6 total frames
mRNA sequences could be in one of three frames on non-transcribed strand
6 total frames
mRNA sequences could be in one of three frames on non-transcribed strand
29
New cards
lactose operon
operon - coordinately controlled set of genes
LacZ
LacY
LacA
LacI
LacZ
LacY
LacA
LacI
30
New cards
LacZ
encodes beta-galactosidase (b-gal)
31
New cards
LacY
encodes lactose permease (LacY)
allows lactose from outside of cell
allows lactose from outside of cell
32
New cards
LacI
encodes lactose repressor (LacR)
control protein of operon
control protein of operon
33
New cards
binding sites in DNA for proteins
aka cis elements
LacO
LacP
LacO
LacP
34
New cards
LacO
lactose operator
binding site for LacR
binding site for LacR
35
New cards
LacP
lactose promoter
binding site for RNAP
binding site for RNAP
36
New cards
LacR
active as tetramer
3 operators (binding sites for LacR)
* LacO1 at +11
* LacO2 +41
* LacO3 -82
3 operators (binding sites for LacR)
* LacO1 at +11
* LacO2 +41
* LacO3 -82
37
New cards
repressing loop
multiple operators lead to formation of repressing loop
excludes RNA from binding lac promoter
excludes RNA from binding lac promoter
38
New cards
lac control mutants
LacI-
LacOc
LacOc
39
New cards
LacI-
mutant LacR cant bind to LacO
no repressor to stop transcription
LacZYA will always be on
no repressor to stop transcription
LacZYA will always be on
40
New cards
LacOc
LacR cant bind mutant LacOc
LacZYA will always be on
LacZYA will always be on
41
New cards
glucose levels as regulators of Lac operon
LacZYA is on only when there is no glucose and only lactose is present
1. when glucose is low, cAMP is high
2. cAMP binds CRP proteins
3. CRP binds control region of lac operon and recruits RNAP
1. when glucose is low, cAMP is high
2. cAMP binds CRP proteins
3. CRP binds control region of lac operon and recruits RNAP
42
New cards
Lac operon control region
\-10 consensus seq: TATAAT
\-10 LacP: TATGTT
\
\-10 LacP: TATGTT
\
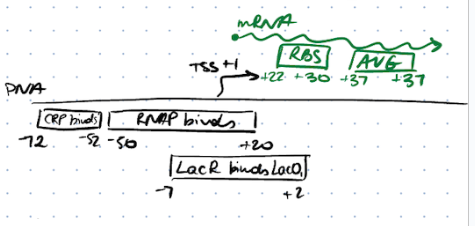
43
New cards
LacP mutagenesis
\-10 sequence
TATGTT → TATATT (higher levels of mRNA)
TATGTT → TAGGTT (lower levels of mRNA)
TATGTT → TATATT (higher levels of mRNA)
TATGTT → TAGGTT (lower levels of mRNA)
44
New cards
assaying b-gal
cell extracts: ONPG (b-gal) → orange color can assay in spectrophotometer
plates: X-gal (b-gal) → blue color on plates
plates: X-gal (b-gal) → blue color on plates
45
New cards
arabinose operon
when AraC bound operon is on
can bind to araO2, araI1, araI2
repressing loop forms in absence of arabinose
araBAD transcribed only in presence of arabinose and lack of glucose
can bind to araO2, araI1, araI2
repressing loop forms in absence of arabinose
araBAD transcribed only in presence of arabinose and lack of glucose
46
New cards
restriction endonucleases
reorganize and cut spec seqs
purified from bacteria
purified from bacteria
47
New cards
EcoRI
restriction endonuclease
5’ overhang
5’-G|AATT|C sequence
5’ overhang
5’-G|AATT|C sequence
48
New cards
KpnI
restriction endonuclease
3’ overhang
5’ G|GTAC|C
3’ overhang
5’ G|GTAC|C
49
New cards
Rsa
restriction endonuclease
blunt end
5’-GT|AC (splits in the middle)
blunt end
5’-GT|AC (splits in the middle)
50
New cards
bacteria consensus sequence
TATAAT (-10 sequence)
51
New cards
eukaryotes consensus seq
TATAAAA (TATA box)
52
New cards
cloning (steps)
\
\
1. making recombinant DNA: cut vector with same restriction enzyme (EcoRI used)
2. mix digested vector and genomic DNAs together in presence of DNA ligase
3. creation of plasmid library
53
New cards
plasmid vectors
must have
* replication origin - plasmid can replicate as cell divides
* selectable marker - only cells that pick up plasmid will grow into a colony
* at least one unique restriction site - where fragments of interest are incorporated into vector
* replication origin - plasmid can replicate as cell divides
* selectable marker - only cells that pick up plasmid will grow into a colony
* at least one unique restriction site - where fragments of interest are incorporated into vector
54
New cards
cDNA library
* collection of vectors, each with a unique DNA sequence derived from unique mRNA
* from mRNA (\~2% of genome) - no introns, promoters, origins
\
* from mRNA (\~2% of genome) - no introns, promoters, origins
\
55
New cards
cDNA library steps
1. isolate polyA+ mRNA from eukaryotic cell
2. add polydT primer (dATP, dCTP, dGTP, dTTP)
3. add reverse transcriptase → makes DNA copy of mRNA template
4. add Rnase (degrades mRNA)
5. DNA folds back on itself and seld primes
6. cut with restriction endonuclease
7. clone DNA fragment into vector
56
New cards
genomic DNA library
derived from genomic DNA (100% of DNA) - includes exons, introns, promoters, origins
57
New cards
base recognizing enzymes
**G**C - Arg409 (argenine)
T**A** - Gln45 (glutaminase)
T**A** - Gln45 (glutaminase)