Lec 16 - RNA Processing
1/54
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
55 Terms
information, chemically, enzymes, life, building, mRNA, Degrading
Outline: RNA Processing
• Like DNA, RNA carries ____
• Like protein, RNA is _____ reactive.
• So, to some extent, RNA can do the jobs of both DNA and protein
• RNA can be used to build machines, including ____.
• RNA may have been the original form of ___ (“the RNA world”).
• RNA processing can be divided into at least three subtopics:
– ____ machines (ribosomal RNA, tRNA, snRNA, snoRNA, riboswitches, miRNA/RNAi, telomerase)
– Packaging and managing information (____)
– __ RNA
ribosomal, translate
rRNA. “_____RNA”. Helps form the ribosome (which is about two-thirds RNA, one-third protein). Ribosomes _____ mRNA and make protein.
transfer, amino acids, mRNA
tRNA. “____ RNA”. Bring ___ __ to the ribosome. Decode mRNA.
coding
mRNA. “messenger RNA”. An RNA copy of the ____ sequence of the gene. Decoded into protein by ribosome
specificity
rRNA, tRNA, and mRNA (and snoRNAs and snRNAs and RNase P RNA) are involved in decoding DNA and making protein, but . . .
• There are many other kinds of RNA. These include miRNA, lncRNA, telomerase RNA, piRNA, hammerhead RNA, guide RNA, riboswitch RNA, and Xist RNA. These have a wide variety of functions, and are not directly involved in decoding. Often, the base-pairing properties of RNAs are used to create ____. See Cech and Steitz 2014.
ribosomal, tRNA, mRNA
Proportions RNA in cells
~75% _____
~20% ______
~5% _____
reactive
RNA is chemically ____
2’OH, stable, enzyme, long, short, mutation
DNA is deoxyribonucleic acid: it does not have the ___, so cannot do this reaction.
DNA is more ____ (good for storing information) but less reactive (bad for being an __).
The high stability of DNA is one reason it is used for ____ genomes, while RNA can be used for some ___ genomes (e.g., RNA viruses such as poliovirus).
Partly because of this stability issue, RNA genes typically have much higher __ rates than RNA genomes
ribozyme
The Reactivity of RNA allows it to promote chemical reactions (an RNA that promotes a reaction is a _____).
intervening sequence
The ______ ____ (IVS) (essentially an intron) of the Tetrahymena ribosomal RNA transcript splices itself out in an all-RNA reaction.
splicing, GTP, proteins, endonuclease
Group I introns promote their own ____ (excision) from a larger RNA.
They require ___, but not protein.
Other than the GTP (a ribonucleotide), this can be an all-RNA reaction.
These introns can encode ______ that assist in splicing, and they can encode homing ____ (which can allow insertion of the “intron” into a DNA strand)
salt, proteins, lariat
Group II Introns
In high ___, self-splicing (i.e., an all-RNA reaction) occurs in vitro.
In vivo, assistance from ______ is required.
Unlike group I introns, reaction does not require GTP.
Reaction involves formation of a ___ , with an A-residue branch-point.
Overall, reaction has considerable similarity to ordinary, protein-mediated splicing.
themselves, eukaryotic, tRNAs
Group I Introns, Group II Introns, and “Ordinary” Introns
Often, genes and their primary RNA transcripts are interrupted by sequences called introns. The introns have to be removed (by “splicing”) to form the mature RNA molecule, that does whatever it does. For instance, it might be an mRNA that is decoded to protein.
There are multiple categories of introns. Group I and Group II introns are special and unusual categories, where the RNA has the remarkable property that, at the RNA level, without involvement of protein, the RNA introns can splice ____ out of the primary transcript.
However, what you will usually hear about are “ordinary” introns, which are found in ____ mRNAs, and are spliced out of the primary transcript by a protein and RNA-containing complex called the spliceosome. These introns are extremely common.
In addition, there are other kinds of introns, such as those in _____.
TP53
Human __ gene locus in Ensembl
separated
Evolution of Splicing: Two Theories
1. Introns First. (Splicing evolved as a way to put together pre-existing but ____ functional protein domains, separated by sequences which we now call introns.)
transposable, out, self
Evolution of Splicing: Two Theories
2. Exons First. (Functional exons were there first, but became interrupted by a ___ element, a parasite that could insert itself into other sequences, but could also splice itself __. This would originally have been a __-splicing (i.e., type I or II) intron.)
useful
Even if the self-splicing parasite theory is correct, in some cases organisms would find uses for the insertion. We do see that some introns are __, but this is consistent with either theory
non-splicing
Selfish, ___-_____ RNA inserts into Essential Gene
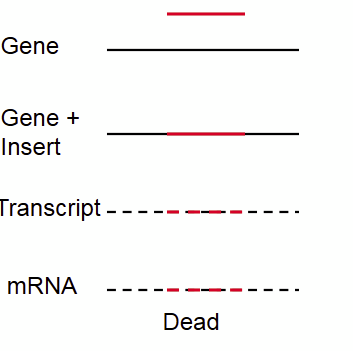
self-splicing
Selfish, ___-_____ RNA inserts into Essential Gene
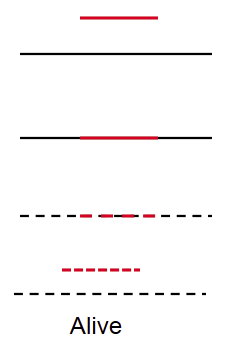
transposition, insertion
The self-splicing RNA can evolve to encode splicing functions, and ___ and ___ functions.
P, introns, enzyme
There are lots of ribozymes
(RNAs that promote reactions)
• RNase __
• Group I and II ___ (not an enzyme)
• Hairpin ribozyme (not an enzyme)
• Hammerhead ribozyme (not an enzyme)
• Peptidyl transferase of 23S rRNA (one of the most frequent and important chemical reactions in life) (this one is an ___)
information, things, both, ribozymes, living, protein
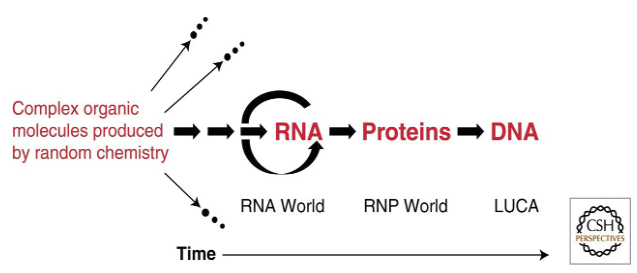
The RNA World
Life requires two things: ability to store ___ (so that it can be passed to the next generation, and evolve), and the ability to do _____. Today, DNA stores information, and protein does stuff, and RNA bridges between them. But RNA can do ____: it can store information, and catalyze reactions.
W. Gilbert proposed that RNA was the original living molecule. RNA could function as the hereditary material (as DNA usually does today, but to some extent also RNA), but also as the structural and catalytic molecules (as RNA and protein do today). Thus all the functions of life could be available in one kind of molecule, or even exactly one molecule. ____ may be living fossils of the RNA world.
Today, in vitro evolution of ribozymes into “__ molecules” (Jack Szostak, Gerald Joyce) is an active area of research.
Present world: DNA (stores info) to RNA to _____ (does things).
Organisms with RNA genomes?
world
An RNA ___ model for the successive appearance of RNA, proteins, and DNA during
the evolution of life on Earth
23S
The ___ ribosomal RNA catalyzes peptide bond formation
ribosomal, peptide, protein
23S _____ RNA catalyzes peptide bond formation
The fact that ____ bond formation is catalyzed directly by RNA supports the idea that our current “DNA-RNA-Protein World” evolved from an “RNA World”.
• Noller paper.
• All three major kinds of RNA (rRNA, tRNA, mRNA) collaborate to make _____—perhaps a sign that RNA is primary.
RNA
A ribosome is a large machine, mostly ___, that makes protein by translating messenger RNA
4, proteins, RNA
RNA Processing to build Ribosomes.
• Eukaryotic ribosomes contain __ large RNA molecules, and many (~60) small __. By mass, the ribosome is about two-thirds __
ribosomal, tRNA, small, messenger
In a growing cell, about 80% of the total RNA is _____ RNA, about 15% is . The remainder is ____ RNAs (snRNA, snoRNA) and ___ RNA.
one, ribosomes, mass
Three of the ribosomal RNAs (18S, 5.8S, 28S) come from __ long transcript, and the mature forms are generated by extensive processing.
In a mammalian cell, there are ~10,000,000 ____. They may constitute 30% of the dry ___ of a bacterial cell.
cutting, trimming, methylation, U, snoRNAs, nucleolus
Processing of the long primary transcript into three mature ribosomal RNAs requires a complicated series of ____ (endonuclease) and ____ (exonuclease) reactions.
In addition, RNA residues are extensively modified, mostly by _____ of the 2’-hydroxyl (the reactive one) of specific riboses, and conversion of specific __ residues to pseudouridine.
The positions of cleavage and the specific sites of 2’-O-methylation and pseudouridylation are determined by about 150 different small nucleolar RNAs (___). These guide reactions by hybridizing briefly to the rRNA molecules (just like the snRNAs guide splicing by hybridizing briefly to the transcript).
All this processing happens in a compartment of the nucleus called the ____.
ribosomal, processing, non
Many snoRNAs are spliced-out introns of genes encoding ______ proteins or ribosome-______ proteins.
In a few cases, snoRNAs are spliced-out introns of otherwise ___-functional mRNAs.
longer, CCA
RNA processing to build tRNAs
Like ribosomal RNAs, tRNAs are processed from ____ primary transcripts. All or nearly all tRNAs are made after cutting and trimming reactions; addition of the __ amino acid acceptor; splicing (sometimes; blue segment below); and extensive modification of nucleotides. The wobble base in the anti-codon, and the base before the anti-codon, are almost always modified
5’, RNase P, bases, spliced
The __ end of the primary tRNA transcript has to be cleaved off to generate the mature tRNA. Cleavage is by ribonuclease P (_ __). In E. coli, RNaseP consists of a 14 kDA protein and a 377 nucleotide RNA. Under the right salt and Mg conditions, the RNaseP RNA (M1 RNA) is a ribozyme capable of cleaving the 5’ end of the primary tRNA transcript on its own, without any protein.
About 10% of the ___ in tRNAs are chemically modified. There are over 100 different modifications, many of them (a) highly conserved; and (b) chemically bizarre.
Some primary tRNA transcripts are __. The mechanism of splicing is completely different from mRNA splicing.
t6A, codon-anticodon
Example of a highly-conserved tRNA modification:
__ (threonyl carbamoyl adenosine).
From Srinivasan . . .Karzai . . . Sternglanz, 2010.
The “KEOPS” complex of five proteins is conserved in archeabacteria, bacteria, and all eukaryotes.
This complex adds t6A (threonyl carbamoyl added to the tRNA adenosine) to all tRNAs that pair with ANN codons. This aids in __-__ pairing
A37
Primer extension analysis of tRNA Ile (AAU) and Val (UAC). tRNAs isolated from wild type (WT), kae1Δ and sua5Δ mutant strains were subjected to primer extension analysis as described in Materials and methods. End-labeled primers specific to either tRNA Ile (AAU) or Val (UAC) were used for the analysis. Cloverleaf structures of the Ile and Val tRNAs are shown. The specific primers used are complementary to the sequence shown in bold face in the cloverleaf structures. The position of the t6A-modified base (__) in the Ile tRNA, marked by horizontal arrows, was confirmed by running length standards and sequence ladders
ribosome,
Three major processing steps for mRNAs
• 5’ Cap: protection, ___- binding for translation.
• 3’ polyA tail. Protection, transport, translation.
• Splicing. Removal of introns. Some mRNAs do not have introns
functional, coding, introns
mRNA Introns
• Many coding sequences (“exons”) are interrupted by sequences called “introns”.
• The initial RNA transcript includes the introns.
• ___ mRNAs are created by removing the introns by “splicing”.
• In humans, ___ RNA is a bit more than 1% of the genome, and __ are about 25%.
intron, coding
Humans have lots of introns
Human dystrophin, the gene involved in muscular dystrophy, has 79 exons, is ~2,200,000 bases long, and spans ~ 0.1% of the genome. The coding region (i.e., ”Open Reading Frame”, or “ORF”) is about 11,000 bases. So, the gene is about 99.5% ___, and about 0.05% ___.
small, big
____ genomes, few introns.
____ genomes, lots of introns
Yeast
___ genes rarely have introns
reading frame, nucleotides
Note: the intron can be in any ______ ____!!
AAC GGG GTCAACATATTTTAG GAT CAA
or
AAC GG GTCAACATATTTTAG G GAT CAA
or
AAC G GTCAACATATTTTAG GG GAT CAA
and, it makes no difference to the
encoded protein if the intron
gains or loses a few nucleotides.
huge
Human genes have lots of __ introns
First part of the human dystrophin (DMD) gene, drawn almost to scale, with the first 26 (of 79) exons. Red arrows show exons (tiny green boxes) everything else is intron.
7 methylguanosine, 5’, 5’
5’ Cap
The 5’ end of the mRNA is modified co-transcriptionally by addition of a __ ____ 5’ Cap. Note that this is added with “backwards” polarity—the __ side of the cap is attached to the __ end of the mRNA via a 5’ to 5’ triphosphate.
initation, exonucleases, cytoplasm
5’ Ccap
The cap has at least three roles. First, it is recognized by proteins for ___ of translation (e.g., eIF4E), which then bring in the ribosome. The cap directs the ribosome to the 5’ end of the mRNA, and is essential for efficient translation. Second, the cap protects the 5’ end of the mRNA from RNA ____. Third, the cap is important for directing the export of the capped mRNA to the _____
5’, 100-150, PAB
3’ end formation
In eukaryotes, the __ end of the mRNA is where the RNA polymerase II starts transcribing.
But the 3’ end is NOT where polymerase stops.
Instead, RNA pol II makes a long primary transcript, which goes past the point that will become the end of the mRNA.
Then, the long primary transcript is cut at the “cleavage and polyadenylation site” to generate the 3’ end.
Then, a “polyA tail” (around __-__ A residues) is added.
Then, a protein called “poly A binding protein” (__) binds to the polyA tail.
site
3′ end mRNA processing: molecular mechanisms and implications for health and disease
Cleavage __ + Poly(A) signal —>Cleavage—>Poly (A) addition
AAUAAA
PolyA site selection is influenced by the presence of a _____ motif (which may involve three motifs), but also by the promoter, distance from the CAP, chromatin, mRNA quality control checkpoints, elongation rate of polymerase....
PAP is Poly A Polymerase. CPSF is Cleavage and Polyadenylation Specificity Factor. CstF is Cleavage Stimulation Factor.
Roles of polyA tail: Attracts poly A binding proteins. Promotes mRNA stability, transport to cytoplasm, translation. PolyA tail must be the right length; too long or too short targets the transcript for degradation via the exosome
exonucleases, poly A tail, 5’ cap, exonuclease
mRNA half-lives
mRNA is protected from 5’ exonucleases by the 5’ cap, and from 3’ ____ by the polyA tail and polyA binding protein.
However, mRNAs have distinct half-lives, ranging from about 2 min to 40 min in yeast (longer in mammals, but still a very wide range). We currently have a very poor understanding of the wide range of half-lives. mRNA turnover comes from attacks from each end, and from cuts in the middle. However, the predominant pathway of turnover is (a) shortening of the __ __ __ via the Ccr4/Not complex; (b) followed by loss of the __ cap; (c) followed by RNA __ digestion from the 5’ end (by the RNA 5’-3’ exonuclease Xrn1).
turnover
Pathways to mRNA _
Poly(A) shorterning
Decapping
5’-3’ nucleic decay
deadenylase
mRNAs might be de-stabilized by signals in the mRNA that attract the ___ complex
translation
Why does shortening of the poly(A) tail at the 3’end of the mRNA lead to decapping at the 5’ end?
One model is that the mRNA forms a loop, in which poly(A) binding protein (PABP) interacts with the ___ initiation complex at the cap. This interaction protects the cap. Only after the interaction is lost (because the poly(A) tail is shortened) can decapping occur. (A. Sachs, A. Jacobson) (Wells, Hillner, Vale, and Sachs 1998.)
exosome, cytoplasmic
RNA Degradation
There are many pathways of RNA degradation, but one important one is the exosome. The ____ is a large complex devoted to RNA degradation, and many kinds of RNA molecules are directed in various ways to the exosome for disposal.
There is a nuclear exosome, for degrading nuclear transcripts, and a ____ exosome, mainly for degrading mRNAs. They differ in that the nuclear exosome has an extra exonuclease, Rrp6
The exosome has a 6-membered ring composed of RNase PH-like proteins, plus three proteins with S1 RNA binding domains, plus two other ribonucleases, Rrp44 and Rrp6.
tag
Poly A and the Exosome
A polyA tail of 70 to 200 nucleotides (depending on the species) stabilizes mRNA; this involves the binding of poly A binding proteins to the tail.
However, other lengths of poly A can be used to __ transcripts for destruction by sending them to the exosome. The TRAMP complex adds short (~15 nucleotide) polyA tails to transcripts using the non-canonical polyA polymerase Trf4 or Trf5, and this directs these transcripts to the exosome. PolyU tails can also serve this purpose.
Recent experiments suggest that abnormally long polyA tails will likewise target transcripts to the exosome. This is a method for controlling abundance of meiotic mRNAs in fission yeast.
Maybe polyA length is measured by wrapping around polyA-binding protein?
6
_-membered ring of the exosome
repress, complementary, circular
Other topics in RNA Processing
• RNA interference and miRNAs. miRNAs are short (22 nuc.), highly-abundant RNAs that hybridize to mRNAs and ___ their expression in various ways.
• Long non-coding RNAs (e.g., Xist, meiRNA).
• Sponge RNAs. Soak up miRNAs by having many copies of a region ____ to the miRNA.
• ___ RNAs. Created by “backwards” splicing. Stable due to lack of ends. Sometimes are sponge RNAs.
• Exon-Junction Complexes. Protein complex deposited on mRNA during splicing. Affects behavior of mRNA.
• Translation Quality Control. Finds mRNAs with defects in translation, destroys them.