Chapter 12 Part One: Regulation Transcription in Eukaryotes
1/86
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
87 Terms
in eukaryotes what regulates transcirption
Tfs
the human genome encodes how many protein encoding genes
20,000
there are many noncoding genes which encode functional ______ that do not get translated into proteins
RNA
what is an example of a protein that is abundant in specialized cells in which they function and cannot be detected elsewhere
hemoglobin can only be detected in red blood cells.
what are the 2 components of a cell that allows it to determine which genes will be trasncribed
short stretches of DNA of defined sequence
gene regulatory proteins recognize and bind to these shrot segments
what are the two components of eukaryotic gene regulation
transcriptional gene regulator
post trasncriptional gene regulation including miRNAs, lncRNAs, and siRNAs
what is the ground state of transcriptional regulation in bacteria vs eukayotes
in bacteria the ground state is always on and represesed state is always off.
In eukaryotes the ground state is always off and the active state is always on
what is the 3 main differences between transcription with bacteria vs eukaryotic orgnaisms
in bacteria all genes get transcribed into RNA by 1 RNA polymerase while in eukayotes ir 3 RNA polymerases
RNA transcripts in eukaryotes must be processed meaning that the 5’ to 3’ ends are modified and introns are spliced out
RNA polymerase II is much lager and complex in Eukaryotes
what does RNA pol II do in Eukaryotes
trasncribes mRNA
what is trans acting and what is cis acting in eukaryotes
regulatory proteins can diffuse and are trans acting
regulatory DNA sequences are on the SAME DNA molecule as the gene making it cis acting
access to transcription regulatory seqeunces tends to be restricted in DNA because
DNA is packaged with proteins in the nucleus
what are enhancers
enhancers are regulatory proteins that can bind directly to DNA with the help of trasncription factors
what are proximal enhancers and what are distal enhancers
proximal: close to core promoter
distal: further away from core promoter
general transcription factors can do what
they can directly bind DNA regulatory sequence within core pomroters
what are the names of regulatory proteins that DO NOT bind directly to DNA
coregulators
what are two types of coregualtors
coactivators
corepressors
what are coactivators (coregulators)
what are corepressors (coregualtors)
coactivaters increase the rate of gene transcription, and are recruited by activtor transcription factors.
corepressors: decrease the rate of gene trasncirption and are recruited by repressor transcription factors
what is TFIID and what does it do
TFIID is a coactivator that interacts with transcirption factors and recruits RNA pol II to gene transcription start site
25% of promoters in yeast and humans contain what ?
the TATA box.
point mutations throughout the proximal enhancer and core prooter regions of B globin gene does what to transcribption?
reduces it
what is the galactose-ultilization pathway in yeast
Gal 1, Gal 2, Gal 7 and Gal 10 encodes enzymes that catalyze steps in a biochemical pathway to convert galactose into glucose.
Gal 3 Gal 4 and Gal80 encodes what
they encode proteins that regulate the expression of enzyme genes
which is the key regulator of the GAL gene expression?
Gal 4 protein because it is a sequence specific DNA binding protein
Gal4 transcription factor binds to ____ elements in yeast
UAS: upstream activating sequences
what are the 2 domains of the Gal4 dimer
Activation Domain
DNA binding Domain
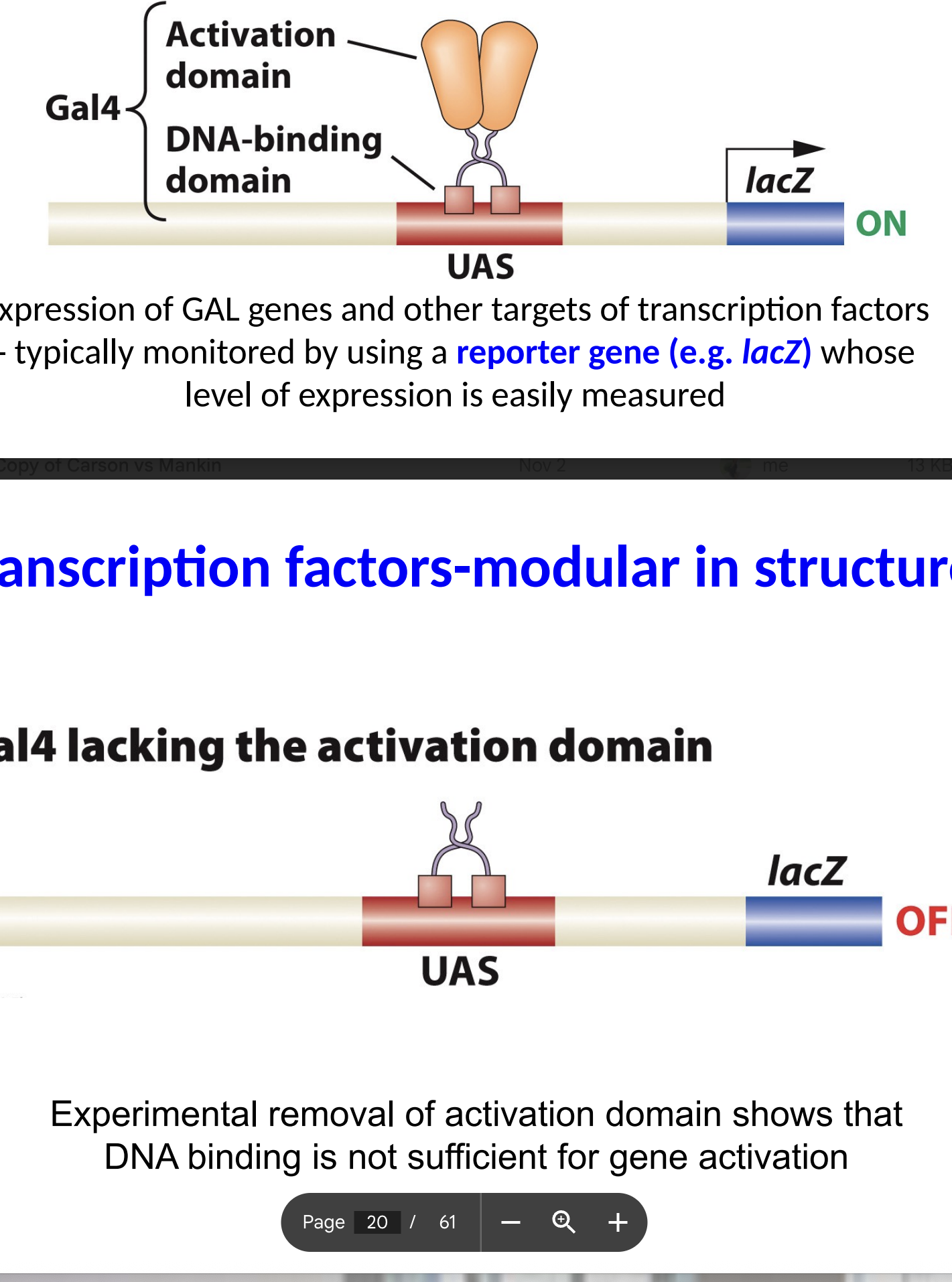
if the Gal 4 does not have the activation domain what happens
LacZ is off
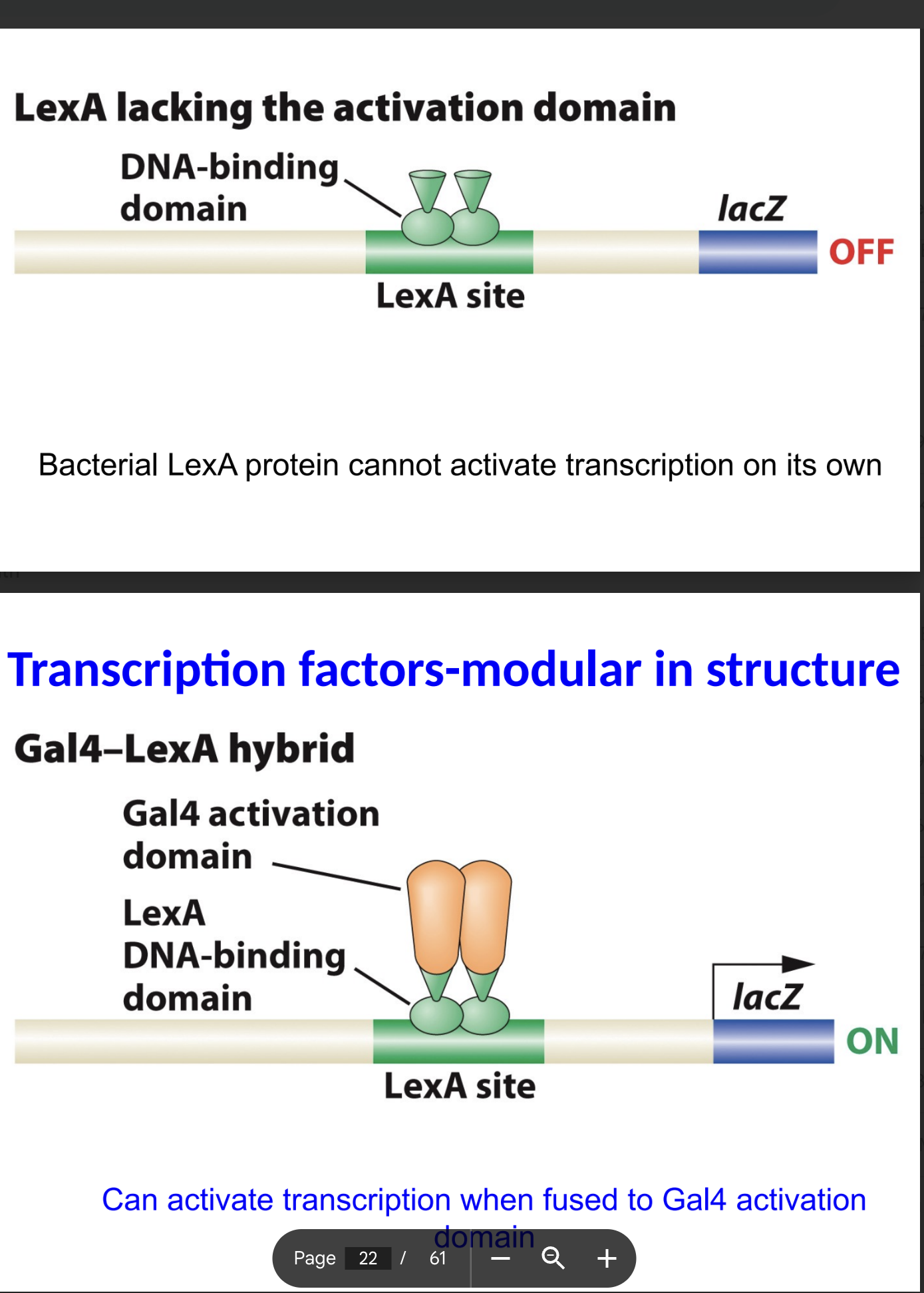
If LexA does not have the activation domian what happpens?
what about when it does have the activation domain?
does not= OFF
does= On
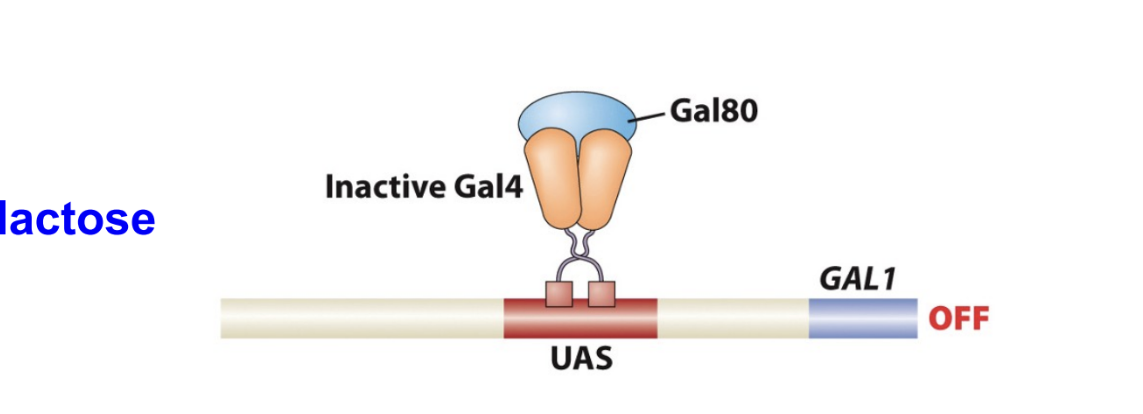
If Gal80 protein is binded to Gal4 what happens
inhibits the Gal 4 activity and the Gal 1 gene is off
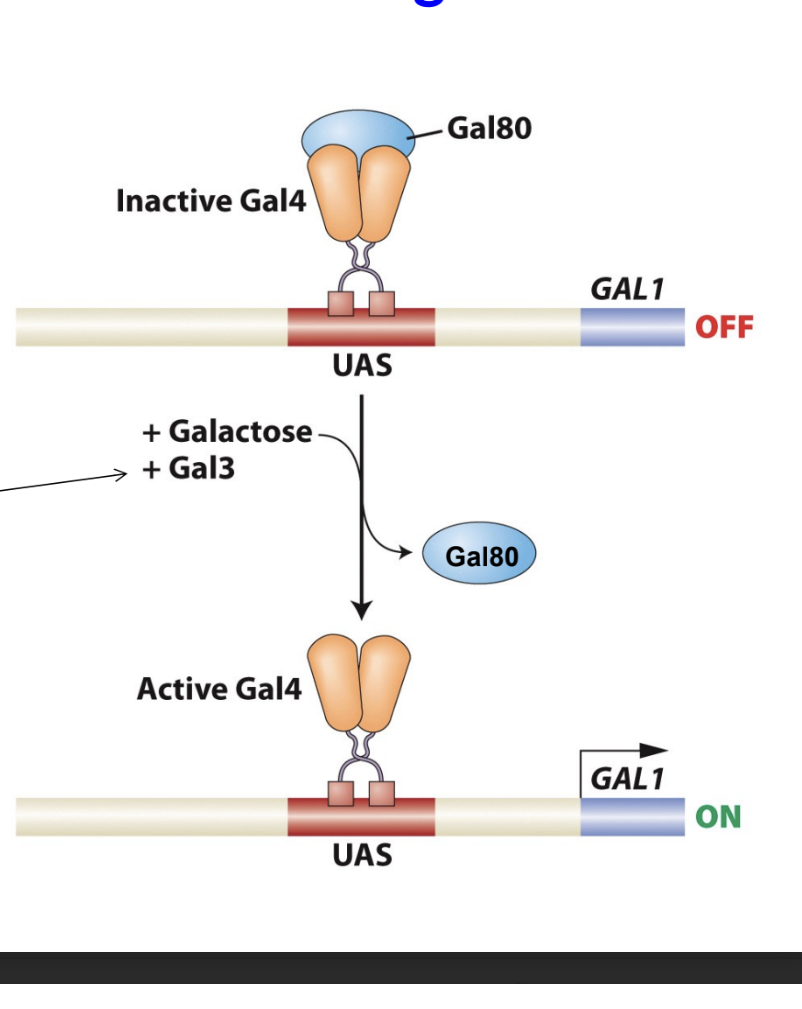
what happens when Gal 3 is added with galactose?
Gal 3 is a sensor and inducer, it will bind to galactose and change the conformation of Gal 80 protein, which causes the Gal80 to release Gal 4, and RNA pol II will activate trasncription turning the gene on
Gal 80, Gal 3 and Gal 4 are all part of a switch state that is determiend by
presence or absense of galactose
the activity of eukaryotic transcriptional regualtory proteins is often controlled by interactions with
other proteins
does Gal4 work in most eukaryotes?
yes it has worked w insects, humans and other species.
what is the first step of transcription initiation in eukaryotes?
eukaryotic promoters are first recongized by general tasncription factors.
what is the function of these GTFs
there role is to attract the core RNA pol II to the so it can begin RNA synthesis at transcription stat sie.
the preinitiation complex also known as the PIC is made up of what
general transcription factors and RNA poll II
the assembly of the PIC complex begins with what
TFIID. this leads to the recurimtent of RNA pol II and other GTFs
why is TFIIH important
it is required to open the transcription bubble and phosphorylate the carboxy terminal domain known as CTD
what does Gal 4 protein bind to in order to recruit RNA pol II to gene promoters. Is this a direct or indirect interaction.
Gal 4 protein must bind to multiple protein complexes, including the TFIID and mediator compleces.
This is indirect
what is the MCM1 Transcription factor
it is expressed in all three cell types, but its function is changed with interactions of alpha1 and alpha2
compare transcriptional regulation with bacteria vs eukaroyes
there are diff ground states in bacteria its on in euakryotes its off. There are more regulatory proteins in eukaryotes. RNA Pol II is larger than Pol I. Compared with eukaryotes, DNA is naked in bacteria.Eukaryotes chromatin strcutrue impacts gene expression.
what makes up chromatin
11nm nucelosomes that fold upon one another into a chromatin fber
a nucelosome contains a protein core made up of _ ______ molecules the core histones
8 histone
Histone H1 is a ______ histone because it binds the DNA that links adjacent nucelosomes
linker
a histone octamer forms a protein_____ around which the double stranded DNA is wound
core
Nucelosomes and chromatin are composed of DNA around _______ histones. The octamer of the histone is made up of two copies of each of the 4 core histones
8
H2A, H2B, H3 and H4)
nucelosomes are spaced at intervals about _____ nucleotide pairs, and they are usually packed together with the aid of histone_____ molecules to form a 30-nm chromatin fiber.
200
H1
which is more compacted euchromatin or heterochromatin
heterchromatin is more compacted
compare euchromatin
heterochromatin
constituitve chromatin
euchromatin: least compact
hetero: most compacted, coated w proteins
consittiutive” remains heterochromatic throughtout cell cycel concentrated at telomeres and centromere
what is the chromatin structure
insulator, enhancer, promoter
nucelosomes cover most regions but are excluded from _____ and ________ of active genes to create ________ _______ _______
enhancer and promoter
nucleosome free regions
in chromosomes, group of genes are kept apart by _______ formed by interaction of insulator binding proteins such as CTCF
TADS— topologically associating domains
what happens with chromsoones with tads
enhancers bound by TFs are positioned to act on GTS at particular promoters
what do insulators do?
divide chromosomes into precisely defined loops that determine which enhancer promoter interactions are allowed and which are not
when there is packaging of eukaryotic DNA into chromatin what does that mean
much of the DNA is not readily accessible to trasncription machinery
eukaryotic genes that are generally inaccessible are
silent unless activated
the modifictiion of chromatin structure is important in (3)
gene regulation
DNA replication
DNA repair
in chromatin modification what happens
enzymes alter te chemical structure of
amino acids in histones OR
nucleotides in DNA
what are the most conserved proteins ? meaning they are near dientical in all eukaryotic organsimss
histones
definiton of nucelosoems
DNA wrapped around 8 hisotones octamer
H2A
H2B
H3
H4
histone tails are subject to what
many different types of post trasnaltional covalent modifications that control aspects of chromatin structure and function
histone proteins with amino terminyl ends make an ________ contact with the phosphate backbone of the DNA
electrostatic
when do covalent modification occur with histone tails
after the histone protein has been translared, and incorporated into the nucelosome
the ________ side chain of the lysine amino acid residues in histones.
how is it neutalzid?
+
it is neutralized with post translational addition of an acetyl group.
what is an acetyl group
CH3-C=O
histone acetylytransferases (HATs) is what ?
histone deacetylases (HDACs) is what?
catalyzation acetylation (the use of acetyl-coA as a donor of the acetyl group ) ADDITIOn
HDACs is the catalyze removal of the acetyl group
Acetlyatio leads to more _____ chromatin by loosening interactins between histones and DNA
open
what happens with Acetylation?
it nuetrlaizes the + charge of the lysine and decreases teh affinity of te lysine for the negatvie charge of the phosphate group of the DNA and reduces the electrostatic interactions between the adjacent nucleosones.
deacetylation _______ chromation compaction and promotes transcription _______.
Acetylation ____ chromation compaction and ______ accessibility of machinery to DNA which promotes trasncription _________
increases
repression
reduces
increases
activation
how does histone acetylation alter chromatin structure and facilitate changes in gene expression?
not only does it decrease chromatin comapction, but with other histone modifications, it influences binding of regulatory proteins to DNA.
can histone deacetylation turn off or on gene transcription?
off
in the presence of glucose, GAl1 transcription _____ by MIG1 protein.
repressed
what does the MIg 1 protein recruit
recruits the TUP 1 repressing complex, which recruits the histone deacetylaase, which turns gene transcirption off.
TUP Complex s a ______ meaning that it faciltiates gene repression but is not itself a DNA binding repressor
corepressor
+ charged amino acid can be covalently modified w the addition of
acetyl groups
methyl groups
How many times can lysine be methylated?
How many times can arginine be methylated?
1,2,3 times
1 , 2 times in symmetric or assymetric config
what are some diff types of histone methylation
lysine
monomethylysine (one methyl group)
dimethylysine (two methyl groups)
trimethyllttsine three methyl groups
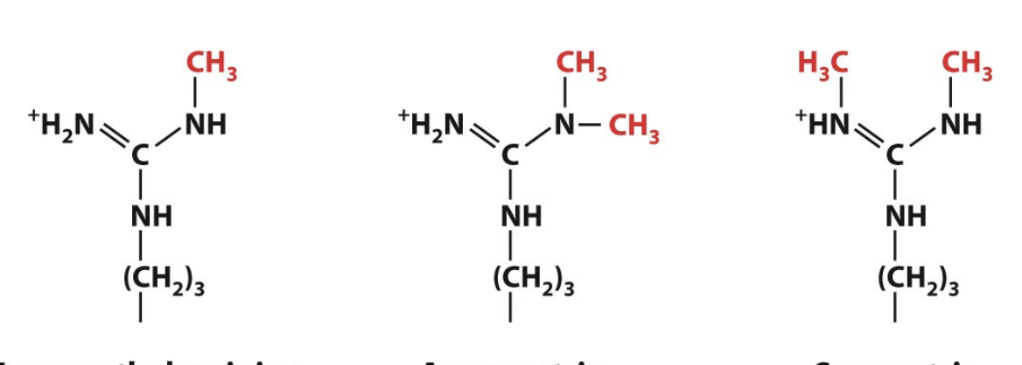
name these three molecules
left is onomethylarginine
middle is assymetric dimethylarginine
right is symmetricdimethylarginine
does the methlyation of lysine residues effect charge?
no but it creates binding sites for other proteins that can either actviate or repress gene expression
Methylation of H3 Lysine Residue for H3K4me3 is associated with ______ of gene expression and is enriched in nucleosomes near start of transcirption
activation
what is post translational modificatiojs and where does it aminly ocur
histone protein tails
what is DNA modification:
additio of methyl groups to DNA resdiues after replication
in verterbrates 85%. of CpGs are methylated. Many of the unmethylated CpGs, are clusted into 200-4000 bp regios called
CpG islands
majority of gene promtoers are asosciated with what
CpG isalnds of unmethylated CpGs
Unmethylated CpG islands at promtoers are generally correlated with _____ chromatin and _____ transcirption.
Methylated CpG are assocaited with ______ chromatin and _______ transcirption
open. active
clossed, repressed
what effects the trasncription of dna
proteins that bind unmethylated or methylated CpGs
what are CpGs
specific sequence of DNA strands where Cytosine nucleotide is immediatly followed by a Guanine nucleotide in the 5’ → 3’ direction. the phosphate group is what links the two nucleotides togehter.
what are the two major mechanisms that operate in eukaryotic cells to enable access of trasncription amchinery to DNA
chromation modificaiton
chromatin remodeling